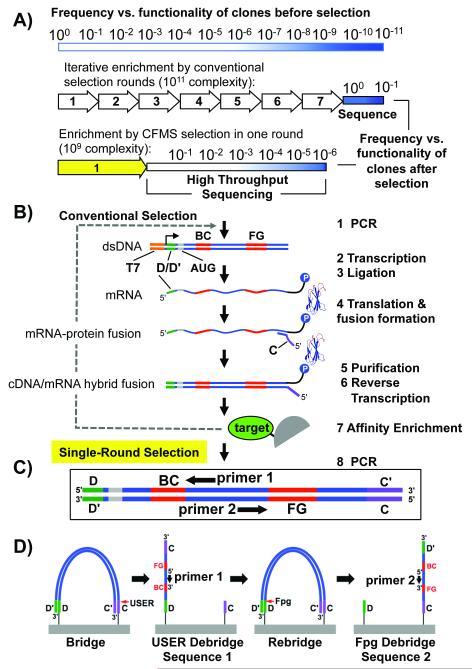
Figure 1.
Selection scheme. A) Graphical representation of functional sequence enrichment by conventional or single round CFMS mRNA display. Functionality, a combination of specificity and affinity, is depicted by a gradient from white (nonfunctional, more common) to dark blue (high functionality, least common). In conventional selection, many rounds of enrichment are performed until most clones are functional. In our single round selection described here, a lower complexity library (~109) combined with improved enrichment efficiency by CFMS (~1000-fold)[5] enables identification of functional clones >1 in 106 by Illumina sequencing. B) Naïve mRNA display library synthesis steps are illustrated (Steps 1-6). The e10Fn3 library was adapted for Illumina sequencing by inserting one of the annealing regions necessary for bridge amplification (“D”) in the 5′ untranslated region. The second chip-annealing/bridge-amplification region (“C”) is added by the reverse transcription primer (step 4). C) For high-throughput selection we identify ligands after one round of selection by sampling the semi-enriched pools through Ilummina HTS (D) using the incorporated annealing regions for bridge amplification and e10Fn3-specific sequencing primers.