Abstract
Genetic variation is usually estimated empirically from statistics based on population gene frequencies, but alternative statistics based on allelic diversity (number of allelic types) can provide complementary information. There is a lack of knowledge, however, on the evolutionary implications attached to allelic-diversity measures, particularly in structured populations. In this article we simulated multiple scenarios of single and structured populations in which a quantitative trait subject to stabilizing selection is adapted to different fitness optima. By forcing a global change in the optima we evaluated which diversity variables are more strongly correlated with both short- and long-term adaptation to the new optima. We found that quantitative genetic variance components for the trait and gene-frequency-diversity measures are generally more strongly correlated with short-term response to selection, whereas allelic-diversity measures are more correlated with long-term and total response to selection. Thus, allelic-diversity variables are better predictors of long-term adaptation than gene-frequency variables. This observation is also extended to unlinked neutral markers as a result of the information they convey on the demographic population history. Diffusion approximations for the allelic-diversity measures in a finite island model under the infinite-allele neutral mutation model are also provided.
Keywords: number of alleles, gene diversity, heterozygosity, response to selection, selection limits, diffusion approximations
THE analysis of the genetic structure of subdivided populations is a key issue in most evolutionary and conservation genetics studies. Genetic variation in subdivided populations is usually estimated as gene diversity (or expected heterozygosity) from gene-frequency data. In addition, genetic differentiation among subpopulations is universally estimated by Wright's (1943, 1969) fixation index (FST), by its multiallelic version (GST, Nei 1973), or by a number of statistics closely related to FST, all of them based on differences in gene frequencies among subpopulations. Moreover, FST or GST, estimated from neutral molecular markers, also provides a reference point for evaluating the strength of divergent selection on quantitative traits (Leinonen et al. 2008; Whitlock 2008).
Allelic-diversity measures, i.e., measures based on the number of different allelic types segregating in the population, are also widely used, particularly in conservation genetics studies. For example, it is recognized that the number of alleles segregating in a population gives basic information regarding past fluctuations in population size (Nei et al. 1975; Luikart et al. 1998). Moreover, the number of rare alleles can be used as an indicator of the amount of gene flow between subpopulations (Slatkin 1985; Barton and Slatkin 1986). In addition, since the number of alleles can be used as an objective conservation criterion, the applications of allelic diversity to conservation issues have been widely investigated (Schoen and Brown 1993; Simianer 2005; Caballero and Rodriguez-Ramilo 2010; Caballero et al. 2010). In this respect, different coefficients of allelic subpopulation differentiation have been proposed for the partition of allelic diversity within and between subpopulations in structured populations (ElMousadik and Petit 1996; Petit et al. 1998; Comps et al. 2001; Foulley and Ollivier 2006; Caballero and Rodriguez-Ramilo 2010). Another differentiation statistic (D) related to allelic diversity was proposed by Jost (2008) for the purpose of estimating differentiation among subpopulations using a partition of genetic diversity in (orthogonal) independent components within and between groups. A substantial debate has been generated recently on whether D should be considered an alternative or a complement for GST (Heller and Siegismund 2009; Jost 2009; Ryman and Leimar 2009; Gerlach et al. 2010; Leng and Zhang 2011; Meirmans and Hedrick 2011; Whitlock 2011; Wang 2012).
The partition of diversity in gene-frequency-diversity or allelic-diversity components leads to rather different conservation strategies (e.g., Caballero et al. 2010), suggesting a complementarity between both types of diversity measures. There is, however, a lack of knowledge about the evolutionary implications of allelic diversity. An aspect on which allelic diversity might have important implications is the response to selection for adaptation toward a changing environment. Whereas short-term response to selection depends on additive genetic variance and, thus, on the expected heterozygosity (Falconer and Mackay 1996), long-term response and selection limits might be more related to the number of alleles initially available for selection. Biallelic locus selection models have shown that the contribution of rare alleles to the selection limit is strongly influenced by the initial population size, so that population bottlenecks restrict the overall response to selection (Robertson 1960; James 1970; Hill and Rasbash 1986). This suggests that the response to long-term selection will increase with the overall number of alleles segregating in the loci controlling the selected trait and could be expected to be larger when more alleles are initially segregating per locus in a set of multiallelic marker loci. Accordingly, in a structured population it may be hypothesized that the long-term rate of adaptation is more dependent on allelic differentiation among subpopulations than on gene-frequency differentiation. Thus, the possibility of a given subpopulation to adapt under a changing environment may depend on the possibility of receiving rare advantageous alleles by migration from other subpopulations (Blanquart and Gandon 2011).
There is also a lack of theoretical predictions for allelic-diversity measures. Ewens (1964, 1972) and Kimura and Crow (1964) provided a simple way to predict the expected number of alleles found in a sample taken from a single undivided population. In the context of structured populations, Tillier and Golding (1988) obtained approximations for the expected number of alleles in samples taken from a single subpopulation that exchanges migrants with other subpopulations. However, these approximations are restricted to very small sample sizes, because the multiple combinations of different allelic types must be taken into account in the calculations. Rannala (1996) derived the distribution of allele frequencies in a sample taken from an island population of fluctuating size, giving a general framework for the analysis of allelic-type frequencies. However, no direct predictive formula for the expected number of alleles per subpopulation was provided in his study.
In this article, we focus our interest on investigating the relationship between adaptive potential and gene-frequency or allelic-diversity measures. We carried out computer simulations of single undivided or structured populations that have reached a mutation–selection–drift equilibrium for a quantitative trait subject to stabilizing selection toward given optima. By changing the selection optima and allowing the population to readapt we investigated the relationship between different gene-frequency and allelic-diversity measures and short- and long-term response to selection. In addition, we developed predictive equations for the within- and between-subpopulation components of allelic diversity by using a diffusion approximation approach in a finite island model with infinite-alleles neutral mutation.
Methods
We first describe the partition of genetic diversity in a structured population through gene-frequency measures or through allelic number measures.
Measures of variation based on gene frequencies
Let us consider a structured population with n subpopulations, where the frequency of allele k for a given locus in subpopulation i is , and KT is the total number of alleles in the whole population. The expected heterozygosity within subpopulations (HS) and the total expected heterozygosity (HT) are
| (1) |
| (2) |
(Nei 1973). The between-subpopulation component of genetic diversity (HT − HS) is also the average Nei´s minimum distance between subpopulations
| (3) |
where is the distance between subpopulations i and j, and the statistic GST (Nei 1973) is
| (4) |
Measures of variation based on allelic numbers
A measure of diversity and differentiation referring to the number of segregating alleles in the population can be made in a way analogous to that for gene-frequency diversity as above (Caballero and Rodriguez-Ramilo 2010). In that approach, ai is the number of alleles present in a random sample of g genes obtained from subpopulation i (ElMousadik and Petit 1996). Its expected value is , where Pik is the probability that allele k is not present in the sample taken from subpopulation i (rarefaction methodology; Sanders 1968; Hurlbert 1971; Kalinowski 2004). When whole subpopulations, instead of samples, are considered, Pik = 0 when the allele is segregating in the subpopulation and 1 otherwise. Then, the within-subpopulation component of allelic diversity is the average allelic number across subpopulations minus one,
| (5) |
The average allelic distance between subpopulations i and j (the average number of alleles present in a given subpopulation that are absent in the other) can be obtained as
(Weitzman 1998; Foulley and Ollivier 2006), and the average distance between all subpopulations is
| (6) |
Hence, a global term (AT) is defined as the sum of both components,
| (7) |
which indicates the average pairwise diversity of subpopulations, i.e., the number of different alleles available in each pairwise grouping of subpopulations, minus 1. Note that AT is not the total number of alleles segregating in the population (KT), but generally a number substantially lower. From the above expressions, a definition of the coefficient of allelic differentiation is
| (8) |
An alternative statistic proposed by Jost (2008) to measure genetic differentiation among subpopulations based on gene frequencies, but highly related to allelic diversity, is
| (9) |
Neglecting, for simplicity, the term n/(n – 1) that corrects for the finite number of subpopulations, and noting that and are the effective numbers of alleles (Kimura and Crow 1964; Crow and Kimura 1970; Jost 2008) of the subpopulations and the total population respectively, D can be expressed as Thus, D is a measure of diversity in terms of effective numbers of alleles.
Studying indicators of adaptive potential in unstructured and subdivided populations
We performed computer simulations with the objective of investigating the extent to which different genetic measures account for the rate of adaptation, both in unstructured and subdivided populations. Simulations were carried out with an in-house C program available on request from the first author. In brief (the detailed procedure is explained below), we considered a quantitative trait under stabilizing selection with a given optimum, in the case of an unstructured population, or with different local optima for each subpopulation, in the case of a structured population. To begin with, we ran different simulations of populations for a wide range of demographic and genetic parameters until an equilibrium was achieved. Each equilibrium provided a different set of initial genetic-diversity measures corresponding to different scenarios. Then, we simulated a shift in the trait’s optima, as could occur due to some change in the environmental conditions, and we tracked the adaptive process (response to selection) as the population mean approached the new optima. We then investigated the relationship between the response to selection that occurred in each simulation and the amount of initial diversity provided by the different genetic-diversity variables.
For the unstructured population scenario, we simulated a population with a constant number N of diploid individuals. A quantitative trait under stabilizing selection was assumed to be controlled by 10 unlinked QTL. In addition, 100 multiallelic neutral unlinked loci (markers) were also considered. New alleles for markers and QTL appeared with Poisson probability at a rate u per generation according to an infinite-alleles model. Each new QTL allele had an additive heterozygous effect on the quantitative trait obtained from an exponential distribution with mean 0.25 environmental standard deviations and a positive or negative sign was assigned with equal probability. The genotypic value of each individual was the sum of the values of their allelic effects and the phenotypic value was obtained by adding to the genotypic value an environmental deviation obtained from a normal distribution with mean zero and variance 1.0. Parents were chosen according to a probability proportional to their fitness, obtained from the function wi = exp[–(Zi – Opt)2/2ω2] (Turelli 1984), where wi is the fitness of individual i, Zi is its phenotypic value, Opt is the optimum value for the quantitative trait (Opt = 0), and ω2 is an inverse measure of the intensity of stabilizing selection. Then, they were mated at random. We assumed ω2 = 25, which represents moderately intense stabilizing selection (Garcia-Dorado and Gonzalez 1996; Mackay 2010).
For the subdivided population scenario, we considered the evolution of the same quantitative trait and genetic markers (simulated according to the procedures described above) for an island model with random migration among subpopulations. In this case, different local optima were considered for the different subpopulations with fixed values (Opt = +5, +4, +3, +2, +1, –1, –2, –3, –4, –5 for the n = 10 subpopulations).
The population (either unstructured or subdivided) was run for 10,000 generations to enable adaptation of the (sub)populations to their optima. After this preadaptation period, a large change in the optima (an increase of +4 environmental standard deviations in the single unstructured population scenario and +10 in the structured population one) was carried out in all subpopulations emulating a global environmental change, but the remainder parameters (subpopulation sizes, mutation and migration rates, intensity of selection, etc.) were assumed to remain invariable. The adaptation of the whole population to the new optima was then evaluated by looking at the mean phenotypic response for the trait after 100 generations.
With the objective of establishing the relationship between the different measures of initial genetic variation and the evolutionary change, simulations in which several parameters were randomly chosen for each run were carried out. Thus, the size (N) of each unstructured population, or of all the subpopulations of each subdivided one, were obtained from a uniform distribution between 100 and 1000. In addition, the mutation rate (u) varied uniformly between 0.00001 and 0.0004 for the unstructured population, while, for the subdivided population, the mutation rate was maintained fixed (u = 0.00001) and the migration rate m = 10–x varied randomly between 0.0001 and 0.1, which was achieved by sampling x from a uniform distribution between 1 and 4. Thus, for a given run, a random combination of N and u (unstructured population scenario) or N and m (structured population scenario) was applied, allowing for a wide range of population genetic parameter values across simulations. Ten sets of 2000 runs were carried out for the single undivided population and five sets of 2000 runs for the structured population scenario. Simulations for the structured population scenario were also run with fixed values of the demographic parameters (N and m).
For each unstructured population, the diversity measures evaluated were the additive genetic variance (VA) for the quantitative trait, the average heterozygosity (H* for the QTL; H for the neutral markers), and the average number of segregating alleles (K* for the QTL; K for the neutral markers). In the subdivided population, the statistics analyzed for the quantitative trait were the within (VW), between-subpopulation (VB), and total (VT) genetic components of the variance and the QST index (QST = VB/[2VW + VB]; Spitze 1993). Furthermore, we computed diversity estimates based on gene frequencies [HS, DG, HT, GST; see expressions (1)–(4)] and estimates of allelic diversity [AS, DA, AT, KT, AST, D; expressions (5)–(9)], for QTL (denoted by an asterisk) or for neutral markers (without asterisk).
Ordinary Pearson correlation coefficients were obtained between the different diversity variables, measured before the change in the optimum, and the short-term response to selection (arbitrarily defined until generation 10; R10), long-term response to selection between generations 10 and 100 (R10–100 or R10–50, and R50–100), and total response during the whole 100 generations period (RT). Multiple linear regression analyses were also carried out using the response to selection as dependent variable and the different genetic-diversity variables as independent ones. All analyses were made with the SPSS package (v. 20).
Evaluation of neutral diffusion predictions for allelic-diversity statistics
To evaluate the precision of the diffusion approximations of allelic variables under a neutral island model, computer simulations were carried out with the C program referred to above assuming a population subdivided into n = 10 subpopulations, each with constant census size N = 1000, where migration among subpopulations occurred under a finite island model, the number of immigrants being Poisson distributed with rate m, i.e., with mean Nm for the number of immigrants per generation and subpopulation. The population was run for 200,000 generations assuming random mating (including random self-fertilization). In each generation, mutation to new allelic variants (infinite-alleles model) for 100 unlinked multiallelic neutral loci occurred with Poisson probability and rate u. The allele types for each marker locus in the last 20,000 generations were used to calculate the population allelic variables (AS, DA, and AT) from Equation 5–7, which were averaged over loci to obtain estimates of AST (Equation 8). The total number of alleles of the population (KT) was also recorded. Simulated values were compared to the corresponding diffusion approximations.
Results
The extent to which the different initial genetic-diversity measures are correlated with the rate of adaptation was investigated by carrying out multiple simulation runs with a range of initial demographic and genetic parameter values corresponding to different effective population sizes and mutation or migration rates, thus implying a substantial variation in responses to selection across runs (see Supporting Information, Figure S1 for an example of selection responses in a particular case). We first present the results for single undivided populations and then for structured populations.
Correlation between diversity measures and response to selection in single undivided populations
To ascertain to what extent each variability measure (quantitative genetic variance, gene-frequency, or allelic-diversity variables) accounts for the response to selection, we carried out a correlation analysis of each variable with response to selection. The variables in this scenario are the initial additive genetic variance (VA), the average initial heterozygosity for the QTL (H*), the average initial number of segregating alleles for the QTL (K*), and the corresponding variables for the markers (H, K). The values of the squared correlation coefficient (R2) with selection response are presented in Figure 1. Figure 1, top, shows that VA is the best predictor of short-term response (R10), whereas the number of alleles (K*) is the best predictor of long-term (R50–100) and total (RT) responses. Diversity for genetic markers (Figure 1, bottom) predicts long-term and total response better than short-term response, correlations being marginally but consistently larger when based on allelic number. However, for all periods, these correlations were much smaller than those for the quantitative trait (QT) or QTL (note the different scale between the top and bottom of Figure 1).
Figure 1.
Squared correlation coefficients (R2) between initial population genetic-diversity variables and response to selection (R10, response to selection until generation 10; R10–50, response from generations 10–50; R50–100, response from generations 50–100; RT, total response until generation 100) for a single undivided population. Population size and mutation rate (u) was varied randomly among simulations (N between 100 and 1000, and u between 0.00001 and 0.0004). VA: initial additive genetic variance. H: initial average expected heterozygosity. K: initial average number of segregating alleles per locus. Terms with an asterisk refer to QTL whereas terms without one refer to neutral markers. The results are based on 10 sets of 2000 simulation runs. Error bars indicate two standard errors of the mean at each side.
Correlation between diversity measures and response to selection in subdivided populations
Constant demographic parameters:
We ran a set of simulations for each of several specific combinations of demographic parameter values (fixed N and m). For each combination of parameters, ordinary correlations were computed between all diversity measures and the short-term (R10), long-term (R10–100), and total response (RT). Table 1 gives the largest ordinary correlations (irrespective of sign) for each combination of parameter values. Regarding neutral genetic marker variables, correlations between diversity variables for genetic markers and response to selection were always very small and nonsignificant, so they are not included in the table. This suggests that when demographic parameters, such as N and m, are invariable, genetic markers do not convey any information on response to selection for a quantitative trait.
Table 1. The largest (irrespective of sign) ordinary correlation between diversity measures and response.
| m | N | Nm | R10 | R10–100 | RT |
|---|---|---|---|---|---|
| 0.0001 | 100 | 0.01 | VW 0.339 | VT 0.171 | VT 0.186 |
| 0.0001 | 500 | 0.05 | VW 0.302 | HT* −0.100 | HT* −0.123 |
| 0.0001 | 1000 | 0.1 | VW 0.272 | VT 0.111 | VW 0.139 |
| 0.001 | 100 | 0.1 | VW 0.347 | VB 0.313 | VT 0.453 |
| 0.001 | 500 | 0.5 | DG* −0.426 | DA* 0.125 | AST* 0.146 |
| 0.001 | 1000 | 1 | VB 0.300 | DA* 0.106 | AST* 0.139 |
| 0.01 | 100 | 1 | VT 0.714 | KT* 0.276 | VT 0.358 |
| 0.01 | 500 | 5 | DG* −0.444 | KT* 0.139 | KT* 0.126 |
| 0.01 | 1000 | 10 | DG* −0.393 | AST* 0.099 | D* –0.149 |
| 0.1 | 100 | 10 | VW 0.378 | AT* 0.307 | AS* 0.394 |
| 0.1 | 500 | 50 | HT* 0.645 | DG* −0.144 | AS* 0.262 |
| 0.1 | 1000 | 100 | HT* 0.709 | D* –0.260 | AT* 0.186 |
The scenario refers to a subdivided population with n = 10 subpopulations of size N, and migration rate m per generation, mutation rate u = 0.00001 and strength of stabilizing selection given by ω2 = 25. Results are based on 2,000 simulations per combination of N and m values. The variables included in the model refer to the quantitative trait and QTL: VW, VB, VT, QST, HS*, DG*, HT*, GST*, AS*, DA*, AT*, AST*, D*, and KT* (see text for definitions). In all cases, these correlations were significantly different from zero with P < 10−5. Allelic-diversity parameters are shown underlined. R10, response to selection until generation 10; R10–100, response from generations 10 to 100; RT, total response until generation 100.
For QT and QTL variables (Table 1), the largest correlations with short-term response (R10) were for different gene-frequency measures or the genetic variance for the trait. This holds for the long-term (R10–100) or total (RT) response in the cases with Nm < 0.5. However, for the cases with Nm > 0.5, the largest correlations mostly involved allelic measures (underlined), suggesting that these convey more information on long-term response than gene-frequency measures in this scenario.
Variable demographic parameters:
We carried out simulations where the values of N and m were randomly changed across runs. The above results with fixed Nm values (Table 1) suggest different outcomes for highly isolated subpopulations (Nm < 0.5, i.e., FST > ∼0.3) or less isolated subpopulations (Nm > 0.5, i.e., FST < ∼0.3). In fact, an inspection to the response to selection achieved for different values of the number of migrants (Nm) per generation and subpopulations (see Figure S2) shows that the two scenarios should be analyzed separately. In the very highly isolated subpopulation scenario (Nm < ∼0.5), an increase in migration implies higher short- and long-term response. In the less isolated subpopulations scenario (Nm > ∼0.5), however, an increase in migration implies higher short-term response but lower long-term response. Thus, in what follows, analyses are made separately for these two levels of migration.
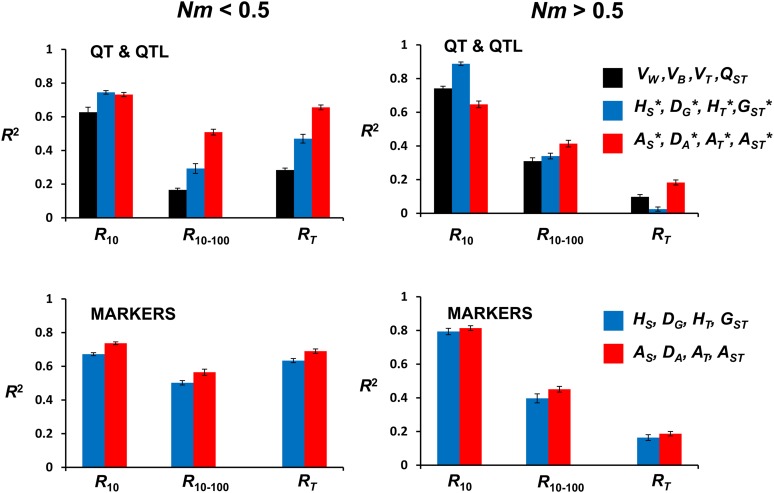
To see which type of variables predicts better the response to selection, we carried out four sets of regression analyses, each including the four main diversity measures. Thus, we performed separate analyses for the quantitative genetic parameters (VW, VB, VT, and QST), the gene-frequency variables for QTL (HS*, DG*, HT*, and GST*), the allelic number variables for QTL (AS*, DA*, AT*, and AST*), and the corresponding sets for marker variables (HS, DG, HT, and GST for gene-frequency variables or AS, DA, AT, and AST for allelic variables). Figure 2 shows the values of R2 for each of these regressions. All five sets of variables explain a relatively large proportion of the variability for selection response (i.e., show large R2 values), except for the total response for the scenario with Nm > 0.5.
Figure 2.
Squared correlation coefficients (R2) between initial population genetic-diversity variables and response to selection (R10, response to selection until generation 10; R10–100, response from generations 10 to 100; RT, total response until generation 100) for a structured population. The scenario refers to a subdivided population with n = 10 subpopulations, Nm migrants per generation and subpopulation, mutation rate u = 0.00001, and strength of stabilizing selection given by ω2 = 25. The variables included in the model are quantitative trait (QT) variables (VW, VB, VT, QST: black bars), gene-frequency variables (HS, DG, HT, GST: blue bars), allelic-diversity variables (AS, DA, AT, AST: red bars). The results are based on five sets of 2000 simulation runs varying the subpopulation size (N) randomly between 100 and 1000 and the migration rate (m) between 0.0001 and 0.1. Error bars indicate two standard errors of the mean at each side.
In contrast with the results obtained for the undivided population scenario or the subdivided population scenario with Nm fixed, measures based on neutral marker loci do not account for less response than those based on QTL. Thus, when demographic parameters are variable, diversity measures based on neutral markers are substantially correlated with response to selection for a quantitative trait. The results also clearly show that allelic-diversity variables (Figure 2, red bars) contain more information about long-term or total response than gene-frequency (blue) or quantitative trait (black) variables.
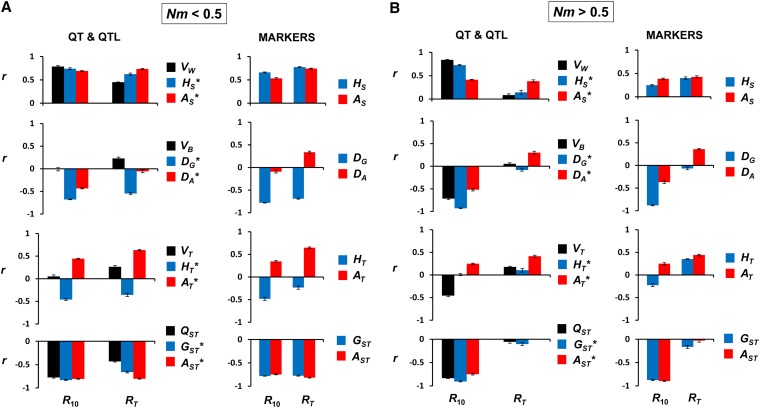
The squared correlations presented in Figure 2 involve five diversity measures. To see more specifically which diversity variables are more correlated with response, Figure 3 gives the correlation for each of these diversity measures with short-term and total response. Correlation coefficients for all variables are presented in Table S1. For the strongly subdivided scenario (Nm < 0.5; Figure 3A), the correlations with the largest magnitude correspond to measures of internal diversity (Figure 3A, top, with r > 0) and of genetic differentiation between subpopulations (Figure 3A, bottom, with r < 0). This holds for all five sets of variables and for both short-term and total response. The measure showing the largest correlation with short-term response is the within-subpopulation additive variance (VW), a correlation that can be ascribed to causality, since short-term response depends directly on this parameter. For total response, however, the best predictors are the allelic measures of genetic differentiation (AST and AST*).
Figure 3.
Ordinary correlation coefficients (r) between initial population genetic-diversity variables and response to selection (R10, response to selection until generation 10; RT, total response until generation 100) for a structured population. The scenario refers to a subdivided population with n = 10 subpopulations, Nm < 0.5 (A), or Nm > 0.5 (B) migrants per generation and subpopulation, mutation rate u = 0.00001, and strength of stabilizing selection given by ω2 = 25. Black bars, quantitative trait variables; blue bars, gene-frequency-diversity variables; and red bars, allelic-diversity variables. Top row: within-subpopulation-diversity measures (VW, HS, AS). Second row: between-subpopulation-diversity variables (VB, DG, DA). Third row: global-diversity measures (VT, HT, AT). Bottom row: differentiation statistics (QST, GST, AST). The results are based on five sets of 2000 simulation runs varying the subpopulation size (N) randomly between 100 and 1000 and the migration rate (m) between 0.0001 and 0.1. Error bars indicate two standard errors of the mean at each side.
For the mild subdivision scenario (Nm > 0.5; Figure 3B), short-term responses are positively correlated with all measures of within subpopulations variability (Figure 3B, top), the largest correlation corresponding to the within-subpopulation additive variance (VW), and negatively correlated with all measures of genetic differentiation (Figure 3B, bottom) or of between-subpopulation genetic distances (VB, DG*, DA*, DG, DA). However, regarding total response, the best predictors are the allelic-diversity variables, both for QT-QTL and markers.
Neutral diffusion predictions of allelic-diversity statistics
In what follows we use diffusion approximations to derive predictions for allelic-diversity measures under an infinite-island neutral model. Let us assume a neutral locus under infinite-allele mutation with rate u per generation, in a population subdivided in n ideal subpopulations of size N, following an island model of migration among subpopulations with rate m. Therefore, the expected gene-frequency differentiation is GST ≈ 1/[1 + M] with M = 4Nm[n/(n – 1)]2 + 4Nu[n/(n – 1)] (Takahata 1983), and the expected effective size of the population (Wright 1943) is
| (10) |
The expected number of alleles whose frequency lies within the range p to p + dp in the equilibrium population is φ(p)dp, where
| (11) |
(Ewens 1964; Kimura and Crow 1964; Crow and Kimura 1970), where θ = 4Neu. Although Equation 11 strictly applies to a random mating population, we show that it provides good approximations regarding several properties of a subdivided population under a wide range of conditions.
Under the infinite-island model, the distribution of allele frequencies within subpopulations (ps) is given by the beta distribution with parameters α = Mp and β = M(1− p), i.e.,
| (12) |
(Wright 1937, 1940), where Γ denotes the gamma function, and p is the whole population allele frequency. Thus, the total number of alleles segregating in the population is
| (13) |
(Ewens 1964, 1972; Crow and Kimura 1970). This latter expectation can also be approximated with a generally lower precision by Ewens (1972) formula,
| (14) |
and by with an even lower precision.
By considering expressions (11) and (12) jointly it is possible to obtain predictions of diversity measures in a subdivided population. This approach has been previously followed by Barton and Slatkin (1986) to obtain the distribution of rare alleles in a subdivided population. The expected number of alleles segregating in each subpopulation is
| (15) |
where
| (16) |
is the cumulative distribution function of between 0 and 1/2N, which gives the probability that a given subpopulation lacks an allele that has frequency p in the overall population. Likewise, the expected number of alleles common to two subpopulations is
| (17) |
The expected allelic diversity within subpopulations (AS) and the expected average allelic difference between subpopulations (DA) are then
| (18) |
| (19) |
Using Equations 7 and 8, this gives which can be computed as
| (20) |
All the above expressions can be modified to account for sampling of g genes within each subpopulation (gn over the whole population). Thus, the total number of segregating alleles in the overall sample of gn copies is
| (21) |
This can also be approximated by Equation 14, replacing 2Nn by gn.
Accordingly, the expected values of KS and KcS would be obtained as above (Equations 15 and 17, respectively) replacing expression (16) with
| (22) |
Precision of the diffusion approximations
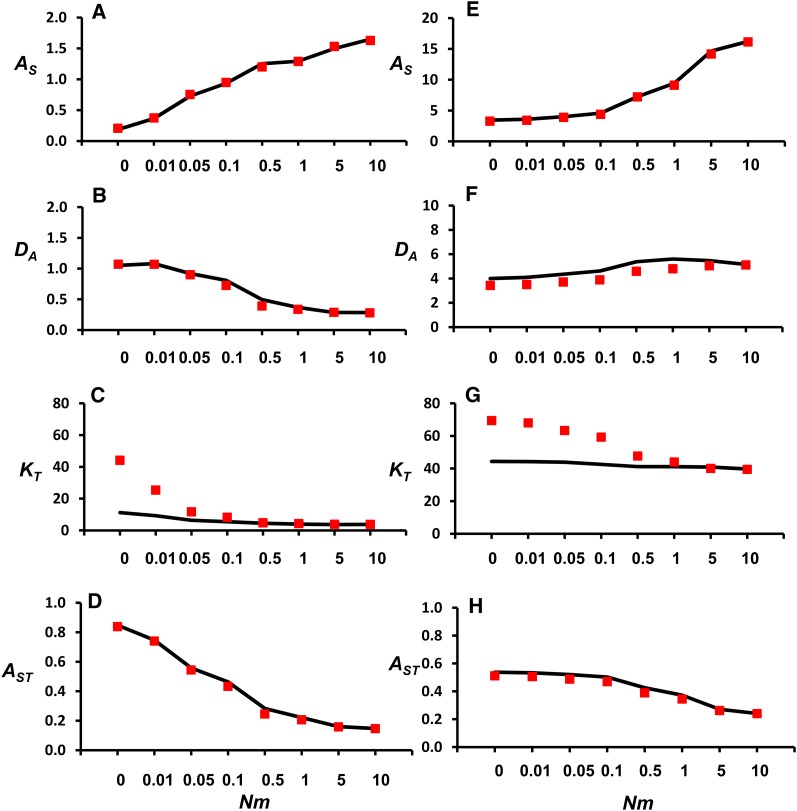
Figure 4 plots predicted and simulated values of the allelic-diversity measures (AS, DA, KT, and AST) against Nm for a range of m values. They are computed for samples of g = 100 neutral genes from each subpopulation for two different mutation rates (a more comprehensive list of results is shown in Table S2 and Table S3). In general, predictions for AS and DA are rather accurate, although those for DA slightly underestimate the simulation values for the large mutation rate scenario. Predictions for KT, however, are well above the values obtained through simulations for low values of Nm. The predictions of AST are very precise in all cases.
Figure 4.
Comparison between computer simulations (lines) and diffusion approximations (symbols) for different allelic-diversity variables. The scenario considered refers to a subdivided population with n = 10 subpopulations, each of size N = 1000 individuals, mutation rate u = 0.00001 (A–D) and 0.0002 (E–H), variable migration rate (m), and g = 100 sampled genes per subpopulation. AS: average allelic diversity within subpopulations. DA: average pairwise allelic distance between subpopulations. KT: average total number of alleles segregating in the whole population. AST: allelic differentiation.
Discussion
The increasing availability of molecular genetic markers for almost any species enables the estimation of variation through gene-frequency diversity (expected heterozygosity) and gene-frequency differentiation (Wright’s fixation index and its derivatives). For multiallelic markers, such as microsatellite loci or allozymes, the number of alleles segregating per locus in the population corrected for sample size is usually also calculated in most genetic-diversity analyses. However, other measures of allelic diversity, such as allelic differentiation among subpopulations are not normally considered. These allelic-diversity estimates can be applied, not only to multiallelic markers but also to biallelic ones, such as SNPs, if different multilocus haplotypes are regarded as alleles (see Pérez-Figueroa et al. 2012 for an example of application).
The issue regarding the distinction between diversity based on “frequencies” and diversity based on “types” has been a topic of general interest in the field of ecology regarding species diversity (Hill 1973; Jost 2007). This distinction has also been discussed in the conservation (Petit et al. 1998; Toro et al. 2009; Caballero and Rodriguez-Ramilo 2010; Caballero et al. 2010) and evolutionary genetics (Jost 2008; Meirmans and Hedrick 2011; Whitlock 2011; Wang 2012) fields. In this article, we have focused on the evolutionary implications of allelic-diversity measures regarding its ability to predict long-term adaptation in single and structured populations. We thus addressed whether the allelic-diversity statistics add something relevant to other genetic-diversity measures in the evolutionary context. Our simulations show that, in fact, allelic-diversity measures are good predictors of long-term response to selection. We have also provided predictions for the allelic-diversity measures for a simple neutral model and start the discussion with this issue.
Theoretical predictions of allelic-diversity measures
Using diffusion approximations under the infinite-allele mutation model we have developed predictive equations for the expected number of alleles within subpopulations in an island model (AS), the differences in allelic types between subpopulations (DA), the allelic differentiation AST, and the total number of alleles in the global population (KT). These are based on the assumption that the expected number of alleles with specified frequencies in the overall population can be approximated using the classical equations (Equation 11) derived for unstructured populations. We have shown that those approximations can be applied rather accurately for structured populations (Figure 4), with the only exception of predicting too many different alleles (KT) when subpopulations are very isolated (Nm < 0.5). The reason is that, for very small Nm, the theoretical prediction of the effective population size (Equation 10) tends to infinity. In this case, the distribution of allele frequencies given by Equation 11 (a formula that assumes panmixia in the population) predicts too many alleles with too small frequencies (often well below the smallest possible value 1/2Nn), which does not match the real distribution for a highly structured population. However, these extremely rare alleles make only a slight contribution to the prediction of the number of alleles in single subpopulations (AS), the allelic differences between pairs of them (DA), or the allelic differentiation index (AST), which are based on the subpopulation gene-frequency distribution (Equation 12). These predictive equations for the number of alleles in subpopulations can be useful, since previous approximations were limited to small sample sizes (Tillier and Golding 1988) or did not focus on obtaining predictive equations for the expected number of alleles (Rannala 1996).
An alternative to the above infinite-allele model that is occasionally used to predict the number of segregating alleles per locus is to assume that each locus contains a virtually infinite number of mutable sites with overall mutation rate u and to compute the expected number of segregating alleles per locus as the expected number of segregating sites (S) per locus. Several studies have been devoted to obtaining predictions of S in structured populations (Tajima 1989; Notohara 1997; Wakeley 1998, 2001). Note that, as θ approaches zero (Nei 1987), Equation 11 for φ(p) approaches the corresponding expression for a model with infinite sites and two alleles per site, which is given by θ/p (1 – p). This implies that, for low mutation rates, the two models (one locus with infinite possible alleles or with infinite sites, each with two alleles) give fairly similar results, but for high mutation rates the number of segregating sites per locus is substantially larger than the number of segregating alleles per locus, because each allele can differ from the rest by more than one site difference. To check this we used expressions (13), (35), and (38) from Wakeley (1998), derived from the time to coalescence, to predict the number of segregating sites per locus in a subpopulation or in the total population in an island model with n = 10, N = 1000, and m = 0.001. Assuming that these numbers were estimated in samples of size g = 100, we obtain (Table S3) that, for u = 0.0002, the values of AS are 9.44 (simulated), 9.11 (diffusion), and 31.30 (Wakeley) and those for KT are 41.14 (simulated), 44.05 (diffusion), and 54.4 (Wakeley), denoting a clear overestimation when using the prediction of segregating sites assuming an infinite-site locus as a proxy for the number of segregating alleles. For the case u = 0.00001, however, both approaches give predictions relatively close to the values obtained by simulation.
Correlation between diversity measures and response to selection
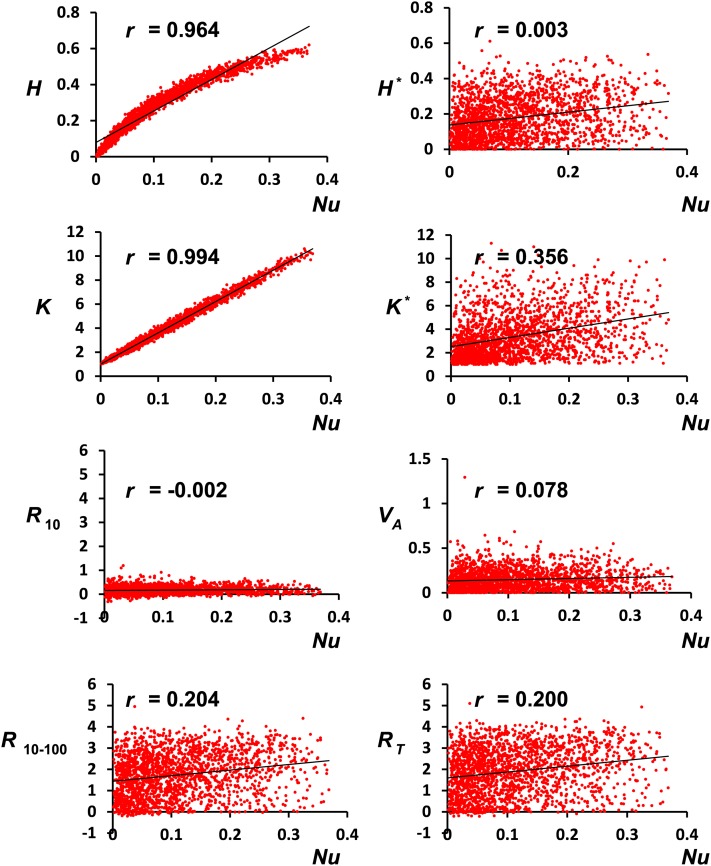
Our results with a single undivided population clearly show that, whereas additive variance is the main factor accounting for the short-term response to selection, as expected from basic theory (Falconer and Mackay 1996), the late and total response are less dependent on the initial additive variance and more strongly correlated with the overall initial number of alleles available for selection (Figure 1). However, to understand the causes of this correlation, it is useful to consider the correlations of response with the diversity measures for genetic markers, which are of course much smaller than those observed for the quantitative trait (QT) and QTL. What is striking in this respect is that the initial number of marker alleles K (or the corresponding initial heterozygosity H) is more strongly correlated with long-term than with short-term response (Figure 1, bottom). This should be ascribed to the information that K or H convey on the number of new mutations that are expected to occur during the adaptive process. The reason is that the expected number of new mutations is proportional to Nu, and K is the best indicator for Nu (r = 0.994), followed by H (r = 0.964). This is illustrated in Figure 5, which also shows, in support of the previous argument, that the initial genetic variance VA and the short-term response scarcely depend on Nu (r = 0.078, r = −0.002, respectively). Similarly, the correlations of VA with K or with H are very small (r = −0.004 or 0.085, respectively). On the contrary, long-term and total response are more dependent on Nu (r = 0.2), because they depend on the future mutational input. Figure 5 also suggests that the larger correlation of late response with K, compared to the corresponding correlation with H, is due to the fact that, in agreement with theoretical expectations, the relationship between H and Nu is not linear, unless Nu values are very small, while the relationship of K with Nu remains linear for a much larger range of Nu values.
Figure 5.
Correlation (r) between several initial population genetic-diversity variables and the product of population size and mutation rate (Nu) for the single undivided population scenario. H: initial average expected heterozygosity for markers. K: initial average number of segregating alleles per locus for markers. H* and K*: corresponding variables for quantitative trait loci (QTL). VA: initial additive genetic variance. R10: response to selection until generation 10; R10–100: response from generations 10 to 100; RT: total response until generation 100. The graphs are based on 2000 simulation runs and the correlations on 20,000 simulation runs.
It is worthwhile to note that the correlation of the initial number of QTL alleles (K*) with response also increases in the long term, although the relative increase is more modest that in the case of K. Furthermore, the correlation of K* with Nu (r = 0.356), although smaller than that estimated for neutral markers, is much larger than the correlation between VA and Nu (r = 0.078). Therefore, it seems reasonable to infer that the reason why the number of QTL alleles is more informative on long-term adaptive potential than the initial additive variance is mainly that it contains more information on the expected mutational input of adaptive variability.
For a subdivided population the situation is more complex, as selection, migration, and drift have a combined impact on local adaptation. On the one hand, when an adaptive equilibrium has been attained, dispersal is generally expected to reduce the level of local adaptation, because the input of suboptimally adapted migrants increases within-subpopulation genetic variance (Lenormand 2002). On the other, for a low migration rate scenario, the process of local adaptation after an environmental change can be enhanced by migration (Blanquart et al. 2012). In fact, when selection fluctuates in time, the level of local adaptation is maximized at intermediate rates of migration (Blanquart and Gandon 2011). Analogously, our simulation results indicate that, for low migration rates (Nm < ∼0.5), increasing migration substantially increases short- and, mainly, long-term response (Figure S2) but, for less subdivided populations (Nm > ∼0.5), increased migration causes an increase of short-term response but smaller late response, having little effect on total response. Thus, we analyzed these two different scenarios independently.
When there is no variation in demographic parameters (fixed N and m), the variability on adaptive potential and genetic differentiation are due to random events that fluctuate through time (including drift and the number of migrations and mutations that occurred). In this situation (see Table 1), the initial values of the genetic measures for QT and QTL are causal indicators of short-term response, but poor indications of long-term adaptation, while those for neutral markers are poor indicators of any response to selection. The results indicate, nevertheless, that allelic-diversity variables correlate more strongly with late and total response for populations that are not heavily structured (Nm > 0.5, i.e., FST smaller than about 0.3), so that allelic-diversity measures become good predictors of adaptation in these scenarios.
The above conclusions generally hold when N and m vary across replicates (Figure 2 and Figure 3). In the strongly subdivided scenario (Nm < 0.5), the best predictors of total response are the allelic measures of genetic differentiation (AST and AST*, with negative correlations). The reason is that, under too strong isolation, subpopulations initially harbor small amounts of genetic variance, and migration provides only small inputs of variability during adaptation, leading to impaired adaptive potential (Figure S2). In the low-isolation scenario (Nm > 0.5), R2 becomes much smaller for long-term or total response than for short-term response since, for this scenario, the metapopulation largely works in the long term like a single undivided population regardless of the m value, so that the information about m contained in these measures becomes irrelevant to long-term response. Even so, all allelic-diversity measures provide a larger correlation with total response than the corresponding variables from gene frequency of QTL or quantitative trait components of variance (see Figure 3B). This supports the relevance of allelic-diversity measures as good indicators of global adaptation.
The large explanatory capacity of measures based on marker loci must be ascribed to the information that they convey on the population structure, rather than to their association with the genetic variance responsible for adaptation. Thus, even if no linkage between loci is assumed, as is the case with our simulations, random variation in migration or any other demographic events will affect simultaneously both QTL and neutral markers, so that neutral markers reflect to some extent the demographic conditions that influence the QTL. Therefore, information from only neutral markers shows an association with adaptive potential that can be similar or even larger than that from QTL. In fact, under variable N and m values, the components of genetic variance for the trait and the diversity measures for the QTL usually contribute less information than the genetic markers in the long term. This probably occurs because QTL are strongly constrained by natural selection, at least with the relatively intense stabilizing selection used in our simulations, so that they convey scarce information on the demographic properties of the population. In addition, the amount of information provided by neutral markers can in principle be enhanced by increasing the number of analyzed markers. Therefore, the results support the use of diversity measures obtained from neutral markers to infer the adaptive potential regarding quantitative traits, at least when an important number of markers can be analyzed.
In summary, we have shown that allelic-diversity measures can be predicted at least for a neutral infinite-alleles island model and that they may contain information regarding the evolutionary potential for adaptation to putative future environmental changes. Our results also imply that the information on long-term adaptive potential contained in diversity measures from marker loci, including those based on allelic types, is due, to a good extent, to the information that these measures contain on Nu and Nm.
Supplementary Material
Acknowledgments
We thank Emilio Rolán-Alvarez, Bill Hill and two anonymous referees for useful comments. This work was funded by Ministerio de Ciencia y Tecnología (CGL2011-25096 and CGL2012-39861-C02), Xunta de Galicia (10PXIB 310044PR and Grupos de Referencia Competitiva, 2010/80), and Fondos Feder: “Unha maneira de facer Europa.”
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Barton N., Slatkin M., 1986. A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 56: 409–415. [DOI] [PubMed] [Google Scholar]
- Blanquart F., Gandon S., 2011. Evolution of migration in a periodically changing environment. Am. Nat. 177: 188–201. [DOI] [PubMed] [Google Scholar]
- Blanquart F., Gandon S., Nuismer S., 2012. The effects of migration and drift on local adaptation to a heterogeneous environment. J. Evol. Biol. 25: 1351–1363. [DOI] [PubMed] [Google Scholar]
- Caballero A., Rodriguez-Ramilo S., 2010. A new method for the partition of allelic diversity within and between subpopulations. Conserv. Genet. 11: 2219–2229. [Google Scholar]
- Caballero A., Rodriguez-Ramilo S., Avila V., Fernandez J., 2010. Management of genetic diversity of subdivided populations in conservation programmes. Conserv. Genet. 11: 409–419. [Google Scholar]
- Comps B., Gomory D., Letouzey J., Thiebaut B., Petit R., 2001. Diverging trends between heterozygosity and allelic richness during postglacial colonization in the European beech. Genetics 157: 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory, Harper & Row, New York. [Google Scholar]
- ElMousadik A., Petit R., 1996. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L) Skeels] endemic to Morocco. Theor. Appl. Genet. 92: 832–839. [DOI] [PubMed] [Google Scholar]
- Ewens W., 1964. Maintenance of alleles by mutation. Genetics 50: 891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W., 1972. Sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3: 87–112. [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Longman, Harlow. [Google Scholar]
- Foulley J., Ollivier L., 2006. Estimating allelic richness and its diversity. Livest. Sci. 101: 150–158. [Google Scholar]
- Garcia-Dorado A., Gonzalez J., 1996. Stabilizing selection detected for bristle number in Drosophila melanogaster. Evolution 50: 1573–1578. [DOI] [PubMed] [Google Scholar]
- Gerlach G., Jueterbock A., Kraemer P., Deppermann J., Harmand P., 2010. Calculations of population differentiation based on G(ST) and D: forget G(ST) but not all of statistics! Mol. Ecol. 19: 3845–3852. [DOI] [PubMed] [Google Scholar]
- Heller R., Siegismund H., 2009. Relationship between three measures of genetic differentiation G(ST), D-EST and G’(ST): how wrong have we been? Mol. Ecol. 18: 2080–2083. [DOI] [PubMed] [Google Scholar]
- Hill M., 1973. Diversity and evenness - unifying notation and its consequences. Ecology 54: 427–432. [Google Scholar]
- Hill W. G., Rasbash J., 1986. Models of long-term artificial selection in finite population. Genet. Res. 48: 41–50. [DOI] [PubMed] [Google Scholar]
- Hurlbert S. H., 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52: 577–586. [DOI] [PubMed] [Google Scholar]
- James J., 1970. Founder effect and response to artificial selection. Genet. Res. 16: 241–250. [DOI] [PubMed] [Google Scholar]
- Jost L., 2007. Partitioning diversity into independent alpha and beta components. Ecology 88: 2427–2439. [DOI] [PubMed] [Google Scholar]
- Jost L., 2008. G(ST) and its relatives do not measure differentiation. Mol. Ecol. 17: 4015–4026. [DOI] [PubMed] [Google Scholar]
- Jost L., 2009. D vs. G(ST): Response to Heller and Siegismund (2009) and Ryman and Leimar (2009). Mol. Ecol. 18: 2088–2091. [Google Scholar]
- Kalinowski S., 2004. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 5: 539–543. [Google Scholar]
- Kimura M., Crow J., 1964. The number of alleles that can be maintained in a finite population. Genetics 49: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen T., O’hara R., Cano J., Merila J., 2008. Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. J. Evol. Biol. 21: 1–17. [DOI] [PubMed] [Google Scholar]
- Leng L., Zhang D., 2011. Measuring population differentiation using G(ST) or D? A simulation study with microsatellite DNA markers under a finite island model and nonequilibrium conditions. Mol. Ecol. 20: 2494–2509. [DOI] [PubMed] [Google Scholar]
- Lenormand T., 2002. Gene flow and the limits to natural selection. Trends Ecol. Evol. 17: 183–189. [Google Scholar]
- Luikart G., Allendorf F., Cornuet J., Sherwin W., 1998. Distortion of allele frequency distributions provides a test for recent population bottlenecks. J. Hered. 89: 238–247. [DOI] [PubMed] [Google Scholar]
- Mackay T., 2010. Mutations and quantitative genetic variation: lessons from Drosophila. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans P., Hedrick P., 2011. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 11: 5–18. [DOI] [PubMed] [Google Scholar]
- Nei M., 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular Evolutionary Genetics, Columbia University Press, New York. [Google Scholar]
- Nei M., Mayurama T., Chakraborty R., 1975. The bottleneck effect and genetic variability in populations. Evolution 29: 1–10. [DOI] [PubMed] [Google Scholar]
- Notohara M., 1997. The number of segregating sites in a sample of DNA sequences from a geographically structured population. J. Math. Biol. 36: 188–200. [DOI] [PubMed] [Google Scholar]
- Pérez-Figueroa, A., S. T. Rodríguez-Ramilo, and A. Caballero, 2012 Analysis and management of subdivided populations with METAPOP, pp. 261–276 in Data Production and Analysis in Population Genomics: Methods and Protocols, chap 15., edited by Pompanon, F., and A. Bonin. Humana Press, Clifton, NJ. [DOI] [PubMed] [Google Scholar]
- Petit R., El Mousadik A., Pons O., 1998. Identifying populations for conservation on the basis of genetic markers. Conserv. Biol. 12: 844–855. [Google Scholar]
- Rannala B., 1996. The sampling theory of neutral alleles in an island population of fluctuating size. Theor. Popul. Biol. 50: 91–104. [DOI] [PubMed] [Google Scholar]
- Robertson A., 1960. A theory of limits in artificial selection. Proc. R. Soc. Lond. B Biol. Sci. 153: 235–249. [Google Scholar]
- Ryman N., Leimar O., 2009. G(ST) is still a useful measure of genetic differentiation - a comment on Jost’s D. Mol. Ecol. 18: 2084–2087. [DOI] [PubMed] [Google Scholar]
- Sanders H. L., 1968. Marine benthic diversity: a comparison study. Am. Nat. 102: 243–282. [Google Scholar]
- Schoen D., Brown A., 1993. Conservation of allelic richness in wild crop relatives is aided by assessment of genetic-markers. Proc. Natl. Acad. Sci. USA 90: 10623–10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simianer H., 2005. Using expected allele number as objective function to design between and within breed conservation of farm animal biodiversity. J. Anim. Breed. Genet. 122: 177–187. [DOI] [PubMed] [Google Scholar]
- Slatkin M., 1985. Rare alleles as indicators of gene flow. Evolution 39: 53–65. [DOI] [PubMed] [Google Scholar]
- Spitze K., 1993. Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135: 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. DNA polymorphism in a subdivided population: The expected number of segregating sites in the two-subpopulation model. Genetics 123: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., 1983. Gene identity and genetic differentiation of populations in the finite island model. Genetics 104: 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier E., Golding G., 1988. A sampling theory of selectively neutral alleles in a subdivided population. Genetics 119: 721–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro M., Fernandez J., Caballero A., 2009. Molecular characterization of breeds and its use in conservation. Livest. Sci. 120: 174–195. [Google Scholar]
- Turelli M., 1984. Heritable genetic-variation via mutation selection balance - lerch zeta meets the abdominal bristle. Theor. Popul. Biol. 25: 138–193. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 1998. Segregating sites in Wright’s island model. Theor. Popul. Biol. 53: 166–174. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2001. The coalescent in an island model of population subdivision with variation among demes. Theor. Popul. Biol. 59: 133–144. [DOI] [PubMed] [Google Scholar]
- Wang J., 2012. On the measures of genetic differentiation among populations. Genet. Res. 94: 275–289. [DOI] [PubMed] [Google Scholar]
- Weitzman M., 1998. The Noah’s Ark Problem. Econometrica 66: 1279–1298. [Google Scholar]
- Whitlock M., 2008. Evolutionary inference from Q(ST). Mol. Ecol. 17: 1885–1896. [DOI] [PubMed] [Google Scholar]
- Whitlock M., 2011. G ’(ST) and D do not replace F-ST. Mol. Ecol. 20: 1083–1091. [DOI] [PubMed] [Google Scholar]
- Wright S., 1937. The distribution of gene frequencies in populations. Proc. Natl. Acad. Sci. USA 23: 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1940. Breeding structure of populations in relation to speciation. Am. Nat. 74: 232–240. [Google Scholar]
- Wright S., 1943. Isolation by distance. Genetics 28: 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1969. Evolution and the Genetics of Populations: The Theory of Gene Frequencies, The University of Chicago Press, Chicago, Illinois. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.