Abstract
Sexual dimorphism at the level of gene expression is common and well documented, but much less is known about how different cis-regulatory alleles interact with the different trans-regulatory environments present in males and females. Here we show that sex-specific effects of cis-regulatory variants are common in Drosophila.
Keywords: gene regulation, trans-regulation, epistasis, imprinting, allele-specific expression
A hallmark of dioecious organisms is sexual dimorphism, phenotypic differences between males and females of a species such as size, coloration, and behavior. Differences in these organism-level exophenotypes are governed by sexual dimorphism in underlying endophenotypes including the regulation of gene expression (reviewed in Williams and Carroll 2009). Gene regulation is central to sexual dimorphism because males and females carry the same genome, except for their sex chromosomes. Indeed, the extent to which the genome is differently expressed in the two sexes is quite striking—estimates in Drosophila suggest that approximately half of the genes in the genome are expressed differently in males and females (Jin et al. 2001; Gnad and Parsch 2006; Innocenti and Morrow 2010).
Mechanistically, the regulation of gene expression is governed by the interaction of cis-regulatory DNA sequences at each gene with trans-regulatory proteins and RNAs present in each cell (reviewed in Wray et al. 2003); the same cis-acting sequences have different activities in the different trans-regulatory environments of males and females. But, do sex-specific differences in the trans-regulatory environment generally have similar effects on alternative cis-regulatory alleles of a gene? Or, put another way, how often do cis-regulatory variants have sex-specific effects? A recent QTL study of expression variation in D. melanogaster found that sex-specific trans-regulatory factors appear to often have different effects on alternative cis-regulatory alleles (Massouras et al. 2012).
Here, we investigate the magnitude of such cis-by-sex effects and compare them to the frequency and magnitude of cis-by-trans effects from other sources. To do this, we used pyrosequencing (Ahmadian et al. 2000) to measure relative allele-specific expression for 11 randomly selected autosomal genes in male and female F1 progeny from reciprocal crosses between the highly inbred Drosophila melanogaster lines zhr and z30 (Begun and Aquadro 1993; Sawamura et al. 1993; Wu et al. 1995; Ferree and Barbash 2009; Coolon et al. 2012). Relative allele-specific expression in heterozygous genotypes provides a direct readout of relative cis-regulatory activity (Cowles et al. 2002; Wittkopp et al. 2004). These reciprocal crosses produced four genetically distinct progeny with identical autosomal genotypes (i.e., heterozygous for the zhr and z30 alleles at all autosomal loci) that differ in the identity of their sex chromosomes and/or the parent of origin for all of their chromosomes (Figure 1A). For each genotype, RNA and genomic DNA were extracted from four biological replicates containing 20 whole flies (7–10 days old) each and analyzed by pyrosequencing using gene-specific primer sets (see supporting information Table S1) and protocols described in Wittkopp (2011).
Figure 1.
Separating the effects of genomic imprinting, epistatic interactions, and sexual dimorphism using reciprocal crosses. (A) Chromosomes present in the parental strains and F1 offspring (excluding the “dot” 4th chromosome) are shown with chromosomes derived from zhr (solid) and chromosomes derived from z30 (shaded). Note that all four types of offspring are heterozygous for all autosomes. (B–G) Six comparisons were performed, contrasting each type of offspring with each other type. For each genotypic type, only the sex chromosomes are shown. The source(s) of interactions potentially affecting relative cis-regulatory activity of autosomal genes in each comparison is shown. Imprinting, genomic imprinting; X, epistatic interactions with variable X- or Y-linked loci; and sex, sexually dimorphic trans-regulatory factors.
Pairwise comparisons among these four genotypes resulted in six tests for differences in relative cis-regulatory activity between alleles of autosomal genes in different trans-regulatory backgrounds (Figure 1, B–F). First, we compared female progeny from reciprocal crosses, which are genetically identical except for any epigenetic marks resulting from the maternal and paternal transmission of alleles known as genomic imprinting (Figure 1B). Next, we compared male progeny from reciprocal crosses, in which relative cis-regulatory activity could differ because of genomic imprinting and/or differences in X and Y chromosome genotypes; genetic differences between the zhr and z30 sex chromosomes have the potential to interact epistatically with cis-regulatory differences between the zhr and z30 alleles of the autosomal genes tested (Figure 1C). In the third and fourth comparisons, we examined male and female progeny from the same cross (Figure 1, D and E). Differences in relative cis-regulatory activity of autosomal genes in these cases could be caused by epistatic effects of trans-acting variants located on the X and/or Y chromosomes and/or differences in the trans-regulatory environment between males and females resulting from sexual dimorphism (i.e., the same pairs of cis-regulatory variants react differently to the trans-regulatory environment of males and females resulting in a sex×cis interaction). Finally, in the fifth and sixth comparisons, we contrasted male progeny from one cross with female progeny from the reciprocal cross (Figure 1, F and G). Differences in relative cis-regulatory activity of autosomal genes in these comparisons could come from genomic imprinting, epistatic effects of genetic differences on the sex chromosomes, and/or sexually dimorphic trans-regulation. In all cases, if relative activity of the zhr and z30 cis-regulatory alleles for autosomal genes is independent of the difference(s) in trans-acting environment, then relative allele-specific expression of these genes should be similar between the two genotypes compared. If, however, the cis- and trans-regulatory differences interact, relative allele-specific expression should differ between genotypes.
Measures of relative cis-regulatory activity (Yijk) were calculated from the pyrosequencing data as log2(zhr/z30) for each gene (i) in each sex (j) from each cross (k), as described in Wittkopp (2011). These data were then fitted to the following linear model using proc MIXED in SAS v10.3 (Cary, NC): Yijk = µ + Sexj (Genei) + Crossk (Sexj (Genei)) + ε. This model controlled for the differences in cis-regulatory activity among genes and allowed us to focus on the effects of different trans-regulatory backgrounds on relative cis-regulatory activity of the autosomal zhr and z30 alleles. We examined the effects of genomic imprinting, epistasis with trans-acting variants on the sex chromosomes, and sex×cis interaction with sexually dimorphic trans-regulatory environments on individual genes using the differences in least-squares means and 95% confidence intervals for these differences derived from this model. An interaction was considered statistically significant for a gene if the 95% confidence interval of the difference did not include zero. This is a conservative test for the absence of an interaction because it does not control for the increased false positive rate resulting from multiple testing.
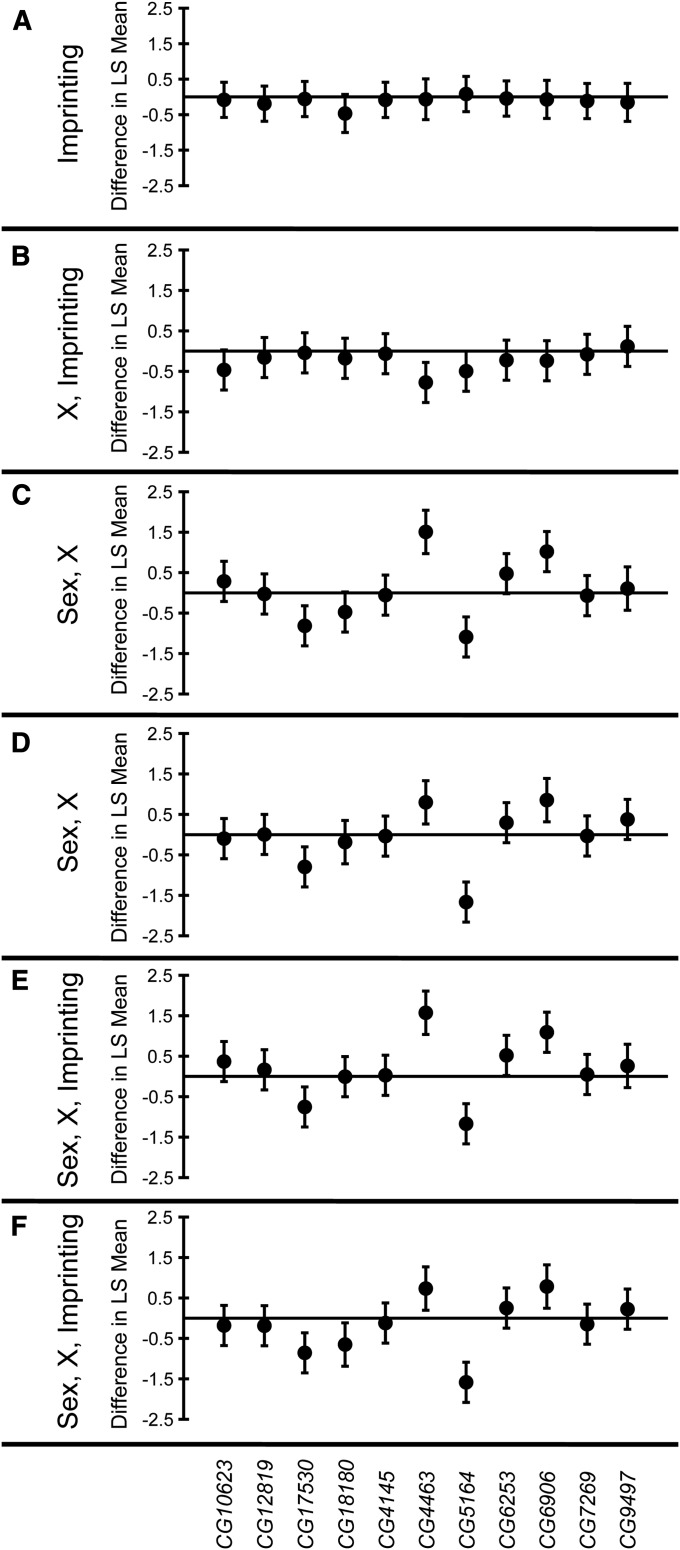
Comparing females from reciprocal crosses (Figure 1B), we found no statistically significant evidence of genomic imprinting for any gene (Figure 2A), consistent with prior studies (Wittkopp et al. 2006; Coolon et al. 2012). In the comparison where relative cis-regulatory activity could be affected by either imprinting or genetic differences between X and/or Y chromosomes (Figure 1C), one gene showed a statistically significant effect (Figure 2B). Given the absence of evidence for imprinting in the first comparison, we conclude that this difference most likely resulted from epistatic effects of one or more trans-acting loci that differ between the zhr and z30 alleles of one or both sex chromosomes. Previous studies provide mixed evidence for this type of epistasis: an intraspecific comparison of D. melanogaster females found no evidence for it among the eight genes tested (Wittkopp et al. 2008), whereas a study of interspecific Drosophila hybrids (D. yakuba and D. santomea) found evidence for it affecting 19 of the 22 genes tested (Llopart 2012). We observed much larger differences in relative cis-regulatory activity in all comparisons between males and females (Figure 1, D–G), with significant differences observed for 6 of 11 genes tested in at least one of the four comparisons (Figure 2, C–F). The statistical significance of the difference in relative cis-regulatory activity varied among comparisons for some genes, but the relative magnitude of the differences was generally consistent among genes in all four comparisons (Figure 2, C–F). This is consistent with differences in trans-regulation between males and females that are similar in all four contrasts and primarily responsible for the differences in relative cis-regulatory activity observed. Statistical significance of the Sexj (Genei) and Crossk (Sexj (Genei)) terms in the full model provide further support for these conclusions (Table 1): after controlling for gene-specific effects, differences between sexes (reflecting sexual dimorphism) explained much more of the total variation in relative cis-regulatory activity (F = 119) than the combined effects of genomic imprinting and epistasis with X- and Y-linked variation captured by the reciprocal crosses (F = 5).
Figure 2.
Relative cis-regulatory activity differed the most between males and females. For each of the six comparisons described in Figure 1, B–G, the difference in relative cis-regulatory activity for each of the 11 genes tested is shown using the least-squares means (LS means) and corresponding 95% confidence intervals derived from the general linear model described in the main text. A–F correspond to B–G in Figure 1, respectively. In each case, the difference was considered to be statistically significant if zero was not contained within the 95% confidence interval. The potential causes of significant differences are indicated for each comparison. Imprinting, genomic imprinting; X, epistatic interactions with variable X- or Y-linked loci; and sex, sexually dimorphic trans-regulatory factors.
Table 1. Summary of effects from the general linear model.
| Effect | d.f. | Sum of squares | Mean square | F | P-value |
|---|---|---|---|---|---|
| Sex(gene) | 21 | 75.82 | 3.61 | 119.23 | <1E-25 |
| Cross(sex(gene)) | 22 | 3.08 | 0.14 | 4.63 | 1.40E-08 |
d.f.. degrees of freedom.
Sexual dimorphism creates differences in gene expression between males and females (Jin et al. 2001; Gnad and Parsch 2006; Innocenti and Morrow 2010), and the data presented here show that these sex-specific trans-regulatory environments often interact differently with alternative cis-regulatory alleles of a gene. This suggests that many cis-regulatory polymorphisms have different effects in males and females. Interactions between sexually dimorphic trans-regulatory environments and species-specific cis-regulatory alleles also were recently observed between D. simulans and D. mauritiana using a different experimental design (Meiklejohn et al. 2013), indicating that these effects are not limited to cis-regulatory variants segregating within a species. Furthermore, while our observations are based on a small subset of the genome, the genes used are not enriched for particular functional groups, chromosomal location, or magnitude of cis-regulatory differences (data not shown), suggesting that the set is unbiased and that sex×cis-regulatory variant interactions are common, consistent with Massouras et al. (2012). These types of interactions can result, for example, from cis-regulatory variants that affect binding sites for trans-regulatory factors that differ between the two sexes (Williams and Carroll 2009; Cooley et al. 2012), as was reported for the Drosophila desatF gene (Shirangi et al. 2009).
Supplementary Material
Acknowledgments
We thank Chung-I Wu and the Bloomington Stock Center for Drosophila strains and Kraig Stevenson and Brian Metzger for statistical assistance. Funding for this work was provided by the National Institutes of Health (5F32GM089009-02 to J.D.C.), the National Science Foundation (MCB-1021398 to P.J.W., 1038099 for the Frontiers Master’s Program support for W.W.), and the Alfred P. Sloan Research Foundation (fellowship to P.J.W.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health, National Science Foundation, or Sloan Foundation.
Footnotes
Communicating editor: J. Birchler
Literature Cited
- Ahmadian A., Gharizadeh B., Gustafsson A. C., Sterky F., Nyren P., et al. , 2000. Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 280: 103–110. [DOI] [PubMed] [Google Scholar]
- Begun D. J., Aquadro C. F., 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550. [DOI] [PubMed] [Google Scholar]
- Cooley A. M., Shefner L., McLaughlin W. N., Stewart E. E., Wittkopp P. J., 2012. The ontogeny of color: developmental origins of divergent pigmentation in Drosophila americana and D. novamexicana. Evol. Dev. 14: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon J. D., Stevenson K. R., McManus C. J., Graveley B. R., Wittkopp P. J., 2012. Genomic imprinting absent in Drosophila melanogaster adult females. Cell Rep. 2: 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles C. R., Hirschhorn J. N., Altshuler D., Lander E. S., 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32: 432–437. [DOI] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A., 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F., Parsch J., 2006. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics 22: 2577–2579. [DOI] [PubMed] [Google Scholar]
- Innocenti P., Morrow E. H., 2010. The sexually antagonistic genes of Drosophila melanogaster. PLoS Biol. 8: e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Riley R. M., Wolfinger R. D., White K. P., Passador-Gurgel G., et al. , 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Llopart A., 2012. The rapid evolution of X-linked male-biased gene expression and the large-X effect in Drosophila yakuba, D. santomea, and their hybrids. Mol. Biol. Evol. 29: 3873–3886. [DOI] [PubMed] [Google Scholar]
- Massouras A., Waszak S. M., Albarca-Aguilera M., Hens K., Holcombe W., et al. , 2012. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 8: e1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Coolon J. D., Hartl D. L., Wittkopp P. J., 2013. The roles of cis- and trans-regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K., Yamamoto M. T., Watanabe T. K., 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics 133: 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi T. R., Dufour H. D., Williams T. M., Carroll S. B., 2009. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 7: e1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. M., Carroll S. B., 2009. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 10: 797–804. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., 2011. Using pyrosequencing to measure allele-specific mRNA abundance and infer the effects of cis- and trans-regulatory differences. Methods Mol. Biol. 772: 297–317. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2006. Parent-of-origin effects on mRNA expression in Drosophila melanogaster not caused by genomic imprinting. Genetics 173: 1817–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2008. Independent effects of cis- and trans-regulatory variation on gene expression in Drosophila melanogaster. Genetics 178: 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray G. A., Hahn M. W., Abouheif E., Balhoff J. P., Pizer M., et al. , 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20: 1377–1419. [DOI] [PubMed] [Google Scholar]
- Wu C. I., Hollocher H., Begun D. J., Aquadro C. F., Xu Y., et al. , 1995. Sexual isolation in Drosophila melanogaster: a possible case of incipient speciation. Proc. Natl. Acad. Sci. USA 92: 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.