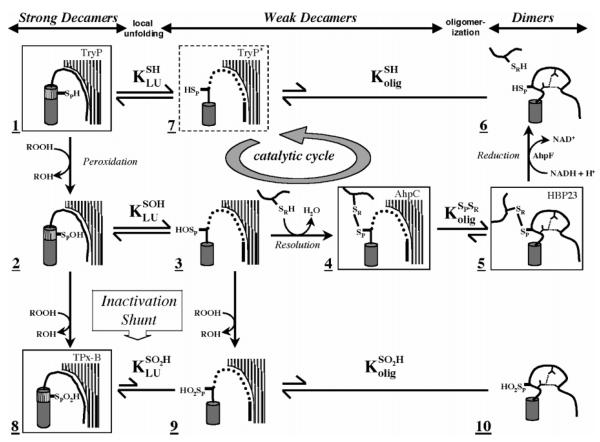
Figure 1.
Structurally detailed model of the 2-Cys peroxiredoxin catalytic cycle and redox-sensitive oligomerization. Each panel represents a different structure of the proposed mechanism. Hatched surfaces at the top right represent the interfacial region (residues 73–84) contributed by an adjacent dimer in the decameric enzyme; panels lacking it indicate dissociation of the decamer to dimers. Thr77 in this interfacial region was targeted for mutagenesis in this study to disfavor decamer formation and potentially affect the catalytic properties of the enzyme. Boxed structures represent determined crystal structures identified by name in the top right corner [TryP (1E2Y), HBP-23 (PrxI, 1QQ2), TPx-B (PrxII, 1QMV), and AhpC (1KYG)]. The dashed box represents an alternate conformation present in the TryP structure. Unboxed structures are proposed intermediates. The active site loop (residues 45–49) containing the peroxidatic cysteine thiol(ate) (SPH), cysteine sulfenic acid (SPOH), or cysteine sulfinic acid (SPO2H) is represented in either its fully folded conformation (hashed cylinder) or its locally unfolded conformation (thick line). The dynamic equilibrium between folded and unfolded states is represented by KLU. Loop residues 40–44 leading up to the active site loop are represented as well-ordered (curved line), loosely packed (dotted line), or restructured (distorted curved line with hydrogen bonds represented by small dashed lines, species 5 and 6). The C-terminus containing the resolving cysteine (SRH or SR) is depicted as a thick line. Redox steps are represented by one-way arrows, and equilibrium steps are denoted with two-way arrows, with the length relative to the proposed direction of the reaction. The catalytic cycle of AhpC is identified by a circular arrow. See the original reference for further discussion (6).