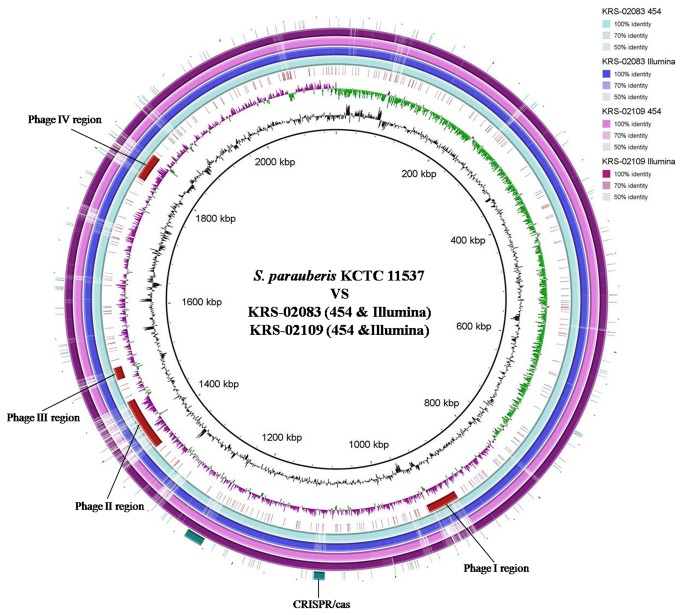
Figure 1. Circular maps from NGS comparing the genomes of S. parauberis strains KRS-02083 and KRS-02109 with that of reference strain KCTC11537.
Beginning with the outermost ring, ring 1 shows the CRISPR/Cas and Tn916 regions encoded in the KRS-20083 genome. Rings 2 and 3 show large contigs from 454 GS-FLX and pair-end reads from the Illumina GA alignment of KRS-02083. Rings 4 and 5 show large contigs from 454 GS-FLX and pair-end reads from Illumina GA alignment in KRS-02109. Ring 6 shows four phage-associated regions in the KRS-02083 genome. Rings 7 and 8 show the GC content and GC skew, respectively, with respect to reference strain KCTC11537. The inner circle shows the scale (bp).