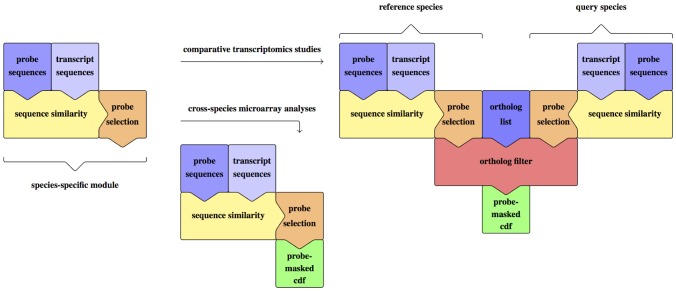
Figure 1. The two possible workflows of the 1 mm approach.
The 1 mm approach can be used in two different ways: For cross-species microarray analyses or for comparative transcriptomics studies. Each of the two workflows results in a probe-masked cdf colored in green. The tips of the colored pieces show the flow of information. The blue colored pieces show the input data provided by the user, whereas the yellow, orange, and red pieces show the two or three steps of the 1 mm approach leading to a probe mask. The species-specific module consists of the sequence similarity step with the microarray-specific probe sequences and the species-specific transcript sequences as input, and the probe selection step that results in a list of probe sets containing only reliable probes. The species-specific module can be used for generating a probe-masked cdf for cross-species microarray analyses of non-model species. Two different species-specific modules can be used with an orthologous gene list for generating a probe-masked cdf for comparative transcriptomics studies.