Table 1. A table of verified candidate genes.
| ae name | locus At | locus Al | probes 1 mm | probes 0 mm | probes gDNA | ΔΔCt | *1 mm | *0 mm | *gDNA | *naive | category |
| 245245_at | AT1G44318 | 314128 | 110-10-0 | -0-0-0 | 11-xxxx-0- | −1.64 | −1.46 | −2.04 | −0.22 | −0.35 | A,B,C,D |
| 245696_at | AT5G04190 | 939816 | 0-0-1-001- | 0-0-00- | 0x0x-00-x- | 3.03 | 1.68 | 2.53 | 0.98 | 0.55 | A,B,C,D |
| 246270_at | AT4G36500 | 490986 | 10110-110 | -0-0-0 | 1-0-x-11- | −2.27 | −2.18 | −2.80 | −1.42 | −1.69 | A,B,C,D |
| 248676_at | AT5G48850 | 494948 | 00-11001-10 | 00-00-0 | -0-1-001x10 | 3.30 | 1.86 | 2.77 | 2.07 | 1.55 | A,B,C,D |
| 251705_at | AT3G56400 | 486080 | 00-10-1-0 | 00-0-0 | 0-x10-1-x- | −2.90 | −2.46 | −2.90 | −1.81 | −1.36 | A,B,C,D |
| 252205_at | AT3G50350 | 485386 | 0-0-0-0-1 | 0-0-0-0- | 0x-x0x0x0x1 | 1.82 | 1.36 | 1.65 | 0.31 | 0.47 | A,B,C,D |
| 252626_at | AT3G44940 | 484892 | -11010-0- | -0-0-0- | -10-0-0- | −1.14 | −0.85 | −0.93 | −1.22 | −0.42 | A,B,C,D |
| 253287_at | AT4G34270 | 491240 | 01100001-00 | 0-0000-00 | 0-100-01-00 | −0.10 | −0.05 | −0.10 | −0.07 | −0.08 | A,B,C,D |
| 253908_at | AT4G27260 | 492072 | 101-0-011 | -0-0-0- | 101x0xx-011 | 3.11 | 2.73 | 2.70 | 2.46 | 2.42 | A,B,C,D |
| 254175_at | AT4G24050 | 492457 | -000100- | -000-00- | -x-00-00xx | −1.14 | −1.01 | −0.95 | −0.60 | −0.64 | A,B,C,D |
| 255788_at | AT2G33310 | 482270 | 1101110-00- | -0-0-00- | 11011-0-00x | 2.46 | 2.09 | 2.22 | 2.01 | 1.94 | A,B,C,D |
| 256131_at | AT1G13600 | 920239 | 1-101-0100 | -0-0-00 | 1-1-1-0-00 | −1.21 | −0.83 | −0.82 | −1.03 | −0.61 | A,B,C,D |
| 257153_at | AT3G27220 | 936451 | 11-00010- | -000-0- | -1xx-00-0-x | −3.95 | −3.80 | −4.21 | −3.65 | −3.24 | A,B,C,D |
| 259407_at | AT1G13320 | 920212 | -000000-000 | -000000-000 | -000000-000 | −0.35 | 0.10 | 0.09 | 0.10 | 0.10 | A,B,C,D |
| 260904_at | AT1G02450 | 470205 | -0-0011110 | -0-00-0 | x-xx-0111- | −2.13 | −1.29 | −1.05 | −0.83 | −0.61 | A,B,C,D |
| 261892_at | AT1G80840 | 477161 | -0000-1100 | -0000-00 | x-0000x-0- | −2.40 | −2.27 | −2.55 | −2.05 | −1.56 | A,B,C,D |
| 263970_at | AT2G42850 | 346095 | 0100001-1-1 | 0-0000- | 0-00-01-1 | −1.67 | −1.11 | −0.82 | −1.16 | −0.97 | A,B,C,D |
| 264867_at | AT1G24150 | 313260 | 010001-1-0- | 0-000-0- | -1-01-0x | −1.37 | −1.63 | −1.85 | −0.92 | −1.27 | A,B,C,D |
| 265452_at | AT2G46510 | 483808 | 001-00- | 00-x-00- | 00-0-xx- | −1.89 | −1.43 | −1.35 | −0.26 | −0.20 | A,B,C,D |
| 265856_at | AT2G42430 | 935111 | 000-010-10 | 000-0-0-0 | -00xx0-0-10 | 1.98 | 1.65 | 2.03 | 0.99 | 0.97 | A,B,C,D |
| 245336_at | AT4G16515 | 493225 | -111-1-11 | – | -1-1-x1xx11 | 2.76 | 2.16 | NA | 0.97 | 0.70 | C,D |
| 245369_at | AT4G15975 | 329916 | -0-1-1- | – | -xx-x- | −2.48 | −1.64 | NA | −0.13 | −0.15 | C,D |
| 245397_at | AT4G14560 | 946923 | -1-110- | – | x1-x-x11-x- | 2.74 | 2.37 | NA | 1.44 | 1.36 | C,D |
| 246993_at | AT5G67450 | 496850 | 0-01-1 | – | 0-x-0-xx- | −2.57 | −0.80 | NA | −1.10 | −0.29 | C,D |
| 247524_at | AT5G61440 | 496303 | -01-01-1 | – | -01-01xx1 | −3.48 | −2.49 | NA | −1.22 | −0.77 | C,D |
| 248858_at | AT5G46630 | 948276 | 1-010- | – | 1-010xx-xx | −0.10 | 0.28 | NA | 0.25 | 0.14 | C,D |
| 250937_at | AT5G03230 | 939701 | 10-110-1 | – | 10-1-0xxx1 | −2.64 | −2.74 | NA | −2.05 | −1.94 | C,D |
| 251910_at | AT3G53810 | 485775 | -1-0-0- | – | x1xxx-xx0- | −1.31 | −1.10 | NA | −0.28 | −0.24 | C,D |
| 253400_at | AT4G32860 | 491410 | -11-010-1 | – | x-11x0-0xx1 | −1.42 | −0.91 | NA | −0.09 | −0.12 | C,D |
| 253959_at | AT4G26410 | 945436 | 1011-01- | – | -11-xxx0- | −0.27 | 0.07 | NA | 0.08 | 0.00 | C,D |
| 261766_at | AT1G15580 | 471758 | -1-0-111- | – | x-1xx0x-x | 4.46 | 1.54 | NA | 0.38 | 0.61 | C,D |
| 262085_at | AT1G56060 | 474673 | -1–100 | – | xxx-x-xx100 | 1.70 | 1.67 | NA | 0.38 | 0.15 | C,D |
| 265256_at | AT2G28390 | 481666 | -1-11100 | – | -xx-11-0- | 0.10 | 0.11 | NA | −0.01 | 0.14 | C,D |
| 266649_at | AT2G25810 | 932757 | 11-01-1- | – | 11-1-x-x | −0.68 | −0.44 | NA | −1.05 | −0.72 | C,D |
| 266820_at | AT2G44940 | 483623 | 1-0-111-11- | – | -0-11-x-1- | −2.24 | −1.44 | NA | −1.73 | −0.87 | C,D |
| 266974_at | AT2G39370 | 482956 | -11-1-1011 | – | -1-x-1011 | 4.02 | 0.85 | NA | 1.56 | 0.67 | C,D |
| 254761_at | AT4G13195 | 333009 | -0-1-0 | – | – | 2.22 | 1.70 | NA | NA | 0.33 | D |
| 265806_at | AT2G18010 | 931672 | 1111-100- | – | – | 3.96 | 0.62 | NA | NA | 0.53 | D |
| 247215_at | AT5G64905 | 951330 | -000- | -000- | – | −4.60 | −1.98 | −1.85 | NA | −0.03 | B,D |
| 248539_at | AT5G50130 | 495070 | -01-00- | -0-00- | – | 2.03 | 1.03 | 1.61 | NA | 0.19 | B,D |
: 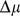 , ae: array element, At: Arabidopsis thaliana, Al: Arabidopsis lyrata.
, ae: array element, At: Arabidopsis thaliana, Al: Arabidopsis lyrata.
For each gene the corresponding array element name and the orthologous gene pair (locus A. thaliana by the TAIR id and locus A. lyrata by the Phytozome gene id) are listed. Additionally, the composition of the probe set in the 1 mm mask, the 0 mm mask, and the gDNA mask are shown. Originally, each probe set consists of 11 probes. The “–” represents a masked probe, “0” a perfectly matching probe, “1” a probe matching with one mismatch, and “x” represents a transcript-unspecific probe. The 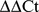 labeled column contains the
labeled column contains the 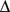 expression values of the 1 h treatment versus no treatment experiments derived from qRT-PCR. The next four columns contain the
expression values of the 1 h treatment versus no treatment experiments derived from qRT-PCR. The next four columns contain the 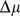 expression values of the 1 h treatment versus no treatment experiments derived from the three probe masking approaches and the non-masking approach. The last column contains the category used for computation of the Pearson correlation coefficient.
expression values of the 1 h treatment versus no treatment experiments derived from the three probe masking approaches and the non-masking approach. The last column contains the category used for computation of the Pearson correlation coefficient.
