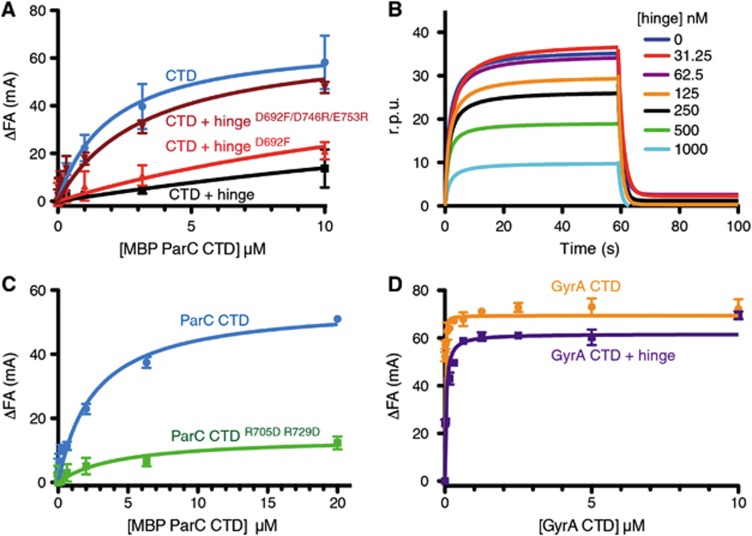
Figure 3.
The hinge prevents the ParC, but not the GyrA CTD from binding DNA. (A) Binding of the MBP-ParC CTD to 20 nM of a fluorescently labelled 20mer duplex oligonucleotide (20 nM), as monitored by relative change (ΔFA) in fluorescence anisotropy (millianisotropy units—mA). Data correspond to titrations with MBP-ParC CTD (CTD) alone, or with 10 μM of the WT MukB hinge, the Asp692Phe mutant, or the Asp692Phe/Asp746Arg/Glu753Arg mutant. See also Supplementary Figure S4. (B) DNA binding by the MBP-tagged CTD as monitored by surface plasmon resonance in the presence of different hinge concentrations. The y axis indicates changes in response units (r.p.u.) due to protein association with the DNA bound sensor chip. The DNA substrate is identical in sequence to the DNA substrate utilized in the fluorescence anisotropy experiments. (C) DNA binding by the MBP-ParC CTDR705D/R729D mutant as determined by fluorescence anisotropy. Substrate is the same as in (A). (D) DNA binding by the tailless E. coli GyrA CTD (aa 531–853) construct as determined by fluorescence anisotropy in the presence and absence of the MukB hinge (10 μM). Substrate is the same as in (A). See also Supplementary Figure S4.