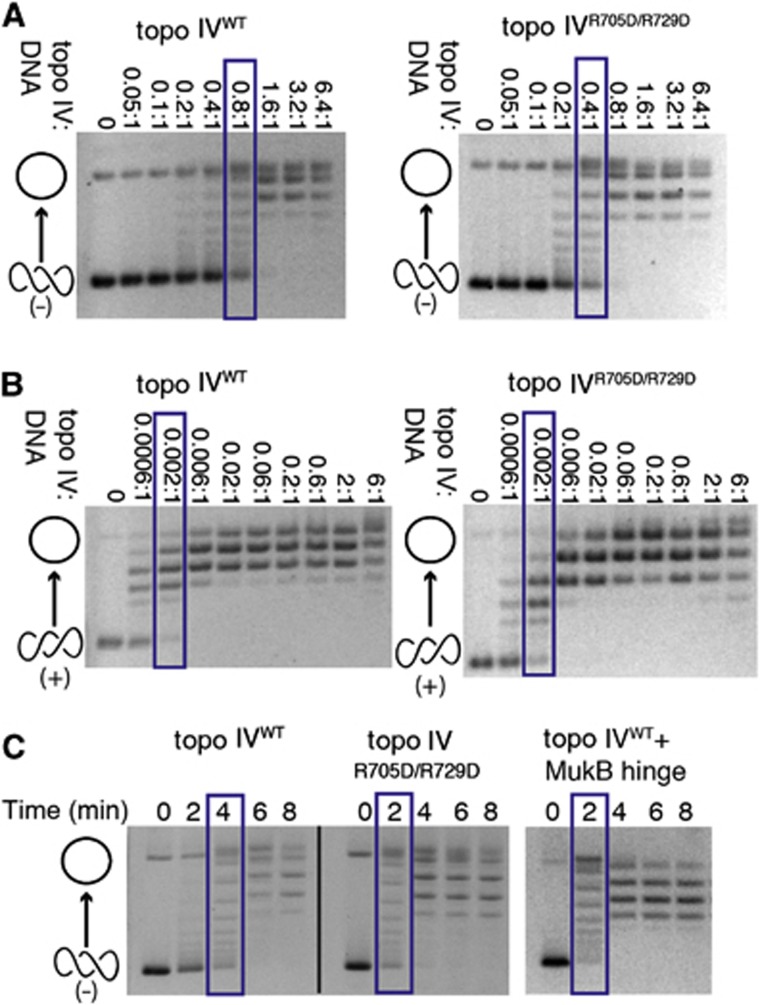
Figure 5.
The hinge masks a DNA-binding site on the CTD that autorepresses the relaxation of negatively supercoiled DNA. Native gels show the relaxation of either negatively or positively supercoiled pSG483 plasmid substrates by WT topo IV or by Arg705Asp/Arg729Asp topo IV (which does not bind MukB). Cartoon representations of topoisomer states are shown on the left side of the gels (open circle—relaxed species and intertwined circles—supercoiled species). Blue boxes indicate protein concentration or time point at which topo IV has relaxed approximately half the supercoiled substrate. Gels are representative of at least three independent replicates from three different protein topo IV preparations. (A) Enzyme titrations showing that mutation of the MukB-binding site on the ParC CTD leads to elevated relaxation activity on negatively supercoiled DNA. The molar ratio of enzyme:DNA is indicated above each gel (reactions contained 7.9 nM supercoiled plasmid). (B) Enzyme titrations showing that mutation of the MukB-binding site on the ParC CTD does not affect the relaxation of positively supercoiled DNA. The molar ratio of enzyme:DNA is indicated above each gel (reactions contained 7.9 nM supercoiled plasmid). (C) Rates of negative-supercoil relaxation are similarly stimulated by either the Arg705Asp/Arg729Asp ParC mutation or binding of the MukB hinge. Reactions contain 500 nM of plasmid DNA and 500 nM of either WT or mutant topo IV (500 nM), together with the presence or absence of 10 μM of the MukB hinge.