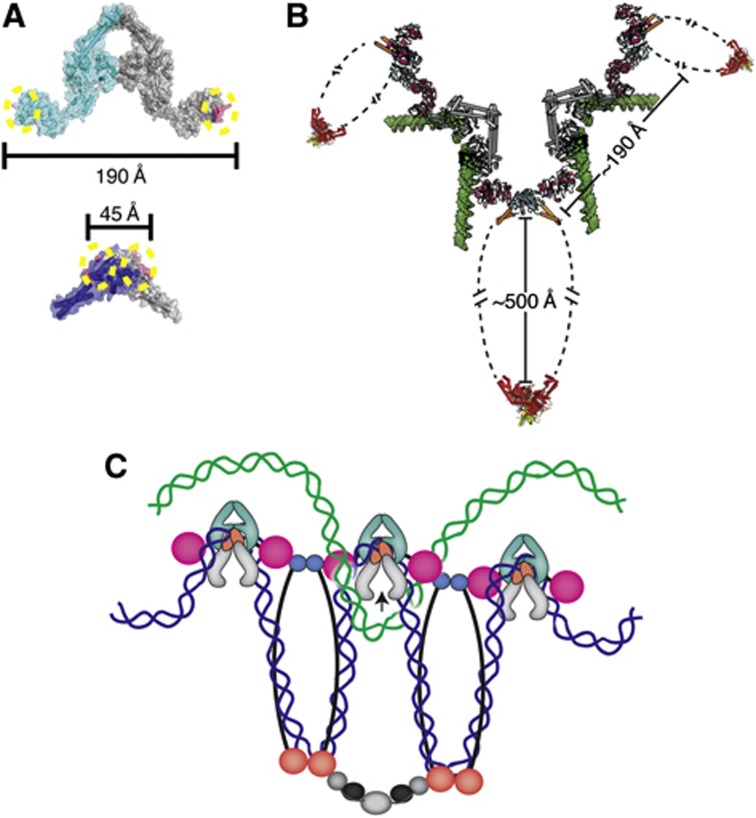
Figure 7.
Physical considerations for the formation of higher order MukB·ParC oligomers. (A) The geometry of the MukB·topo IV interaction does not favour intradimeric complex formation. Upper—MukB-binding sites (magenta with dotted yellow circles) are marked on the crystal structure of the E. coli ParC dimer (cyan and grey) (PDB ID: IZVU). Lower—The ParC-binding sites (magenta with dotted yellow circles) are marked on the crystal structure of the E. coli MukB hinge dimer (navy blue and grey) (PDB ID: 3IBP). The distances between binding sites are shown in Ångströms. (B) The geometry of the MukB·topo IV interaction permits formation of oligomeric arrays. The ParC CTD (pink)·MukB hinge (cyan) coiled-coil arms (orange) structure determined here is shown docked onto with the full-length E. coli ParC crystal structure (grey) (PDB ID: IZVU), modelled with a docked G-segment DNA (green) (superposed from PDB ID: 2RGR). For perspective, the Haemophilus ducreyi MukB head domain (red) in complex with MukF (yellow) (PDB ID: 3EUJ) is also shown. Distances between subunits and domains are labelled. (C) One prospective functional role of a MukB·ParC oligomeric array. Oligomers of MukB and ParC form through the hinge (cyan)·CTD (pink) interaction; MukB oligomers are further stabilized by interactions of their head domains with accessory proteins, MukE (black) and MukF (grey). If MukB and ParC engage the same newly replicated sister chromosome (blue) through DNA bending and looping, then the array predisposes topo IV to remove catenated crossovers formed with the corresponding sister chromosome (green).