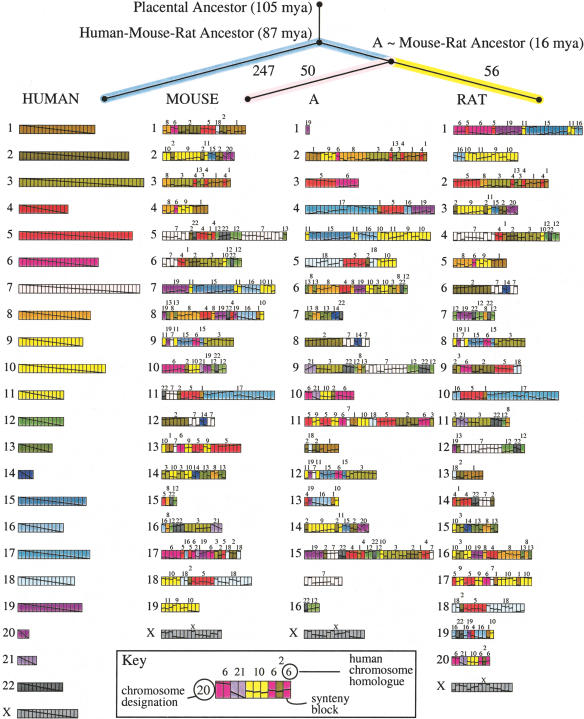
Figure 3.
Ancestral murid rodent genome (A) and evolutionary tree computed by MGR, using mouse and rat with human as an outgroup. Each genome is represented as an arrangement of 391 synteny blocks (longer than 300 kb) as computed by GRIMM-Synteny. The synteny blocks are each represented as one unit, regardless of their length in nucleotides. Chromosomes with too many blocks are split into two lines. Each human chromosome is assigned a unique color, and a diagonal line is drawn through the whole chromosome. In other genomes, this diagonal line indicates the relative order and orientation of the rearranged blocks. The phylogram at the top of the figure indicates the number of rearrangements required to convert each genome (human, mouse, rat) into A, as computed by MGR. The estimated dates of divergence are from Springer et al. (2003).