Fig. 3.
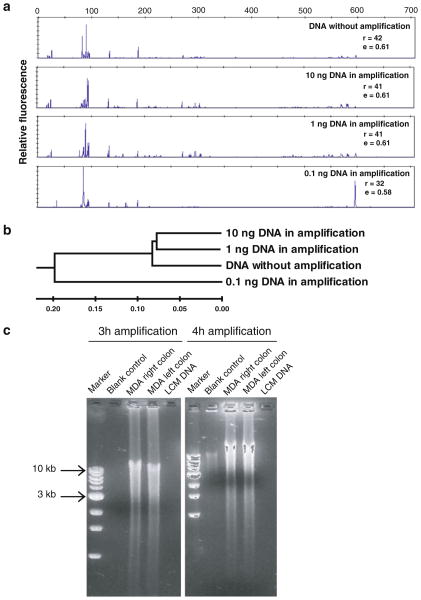
Optimization of phi29 multiple displacement amplification (MDA) with complex DNA templates. a 16S rRNA gene PCR and T-RFLP analysis of fidelity of MDA with the different amount of DNA templates. High fidelity of MDA is observed in the reactions using more than 1 ng DNA as template. Richness (r) and evenness (e) were calculated to show the differences between samples b Dendrogram based on similarity analysis of T-RFLP profiles shows the relationships among samples. The scale bar shows the distance of similarity. c Optimization of Phi29 MDA time on laser capture microdissected DNA samples. Within 3-h amplification, microgram quantity DNA is harvested with minimum endogenous background amplification