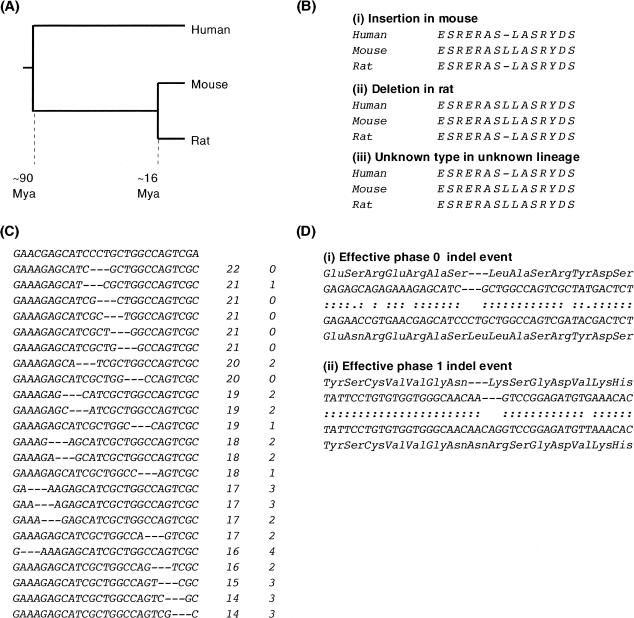
Figure 1.
Identification of coding indel events. (A) The relative phylogenetic relationship of human, mouse, and rat. Mouse and rat last shared a common ancestor ∼16 million years ago (Mya; Springer et al. 2003). Rodents and humans last shared a common ancestor ∼90 Mya (Springer et al. 2003). (B) An alignment gap between mouse and rat could be caused by an insertion in one rodent or a deletion in the other. By alignment to a homologous sequence from an out-group species (here, human) that shared a common ancestor with rodents prior to the divergence of mouse and rat lineages, the mutational event can, through parsimony, be resolved into either an insertion (i) or a deletion (ii) in a specific lineage. (iii) Alignment gaps in the out-group (human) relative to the rodent sequences cannot be resolved through parsimony. (C) Optimal nucleotide alignment. Amino acid alignment indicates the approximate nucleotide position of an insertion or deletion event, in this case a single amino acid deletion in the mouse sequence. The rat sequence (bold) is aligned with the deletion-containing mouse sequence, where a deletion sized gap (1 codon) is allowed to “slide” between –12 and +12 nt of the position indicated by amino acid alignment. These alignments are scored by nucleotide identity between mouse and rat (column 2) and then the number of transition substitutions (column 3). If there are multiple optimal gap positions, the 5′-most position is arbitrarily used for further analysis. (D) Effective phase of indel events. Alignments show mouse (upper rows) and rat (lower rows) amino acid and nucleotide sequences. Amino acids are gapped as indicated by initial amino acid alignment, nucleotides after optimization as shown in panel C. Colons indicate nucleotide identity and periods show transition substitutions. (i) Optimal nucleotide alignment shows that this deletion event is likely to have deleted the third nucleotide of a serine codon and the first two nucleotides of a leucine codon. It is therefore a phase 2 event. However, because the event did not cause an amino acid substitution as well as a deletion, it can be considered an effective phase 0 event (the same consequence on the encoded protein as a phase 0 event). (ii) Optimal nucleotide alignment indicates that this insertion event is also a phase 2 event. In this case, the indel results both in the insertion of an amino acid and the substitution of an amino acid. This can be considered an effective phase 1 event.