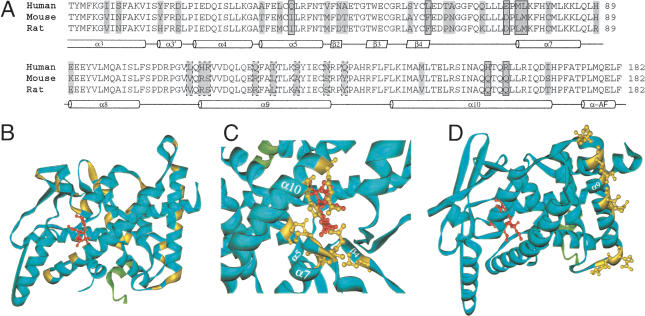
Figure 4.
Variable sites in the LBD of PXR. (A) The variable sites, highlighted in gray, in the protein sequence alignment of the LBDs of the human, mouse, and rat PXRs. The corresponding secondary structure is indicated below the sequence alignment: the α-helix is represented by the cylinder, and the β-sheet by the parallelogram. The seven variable sites at which the amino acid residues line the ligand-binding pocket and the ones found in the α-helix 9 and its vicinity are boxed with the solid line and broken line, respectively. Other available LBD sequences of the rhesus, pig, rabbit, dog, chicken, and zebrafish PXRs are omitted from the sequence alignment presented here, because the inclusion of them does not introduce changes to the general variation pattern. (B) The same sites, highlighted in yellow, in the tertiary structure of the LBD of the human PXR (the blue solid ribbon). The agonist is shown as the small red molecular structure bound in the receptor's ligand-binding pocket, and the coactivator in fragment is shown as the green solid ribbon. Details of the variable sites, with the side chains of the amino acid residues at these sites shown in yellow, in the ligand-binding pocket (C) and in the α-helix 9 and its vicinity (D) are shown.