Abstract
7.87 to 10.5 eV vacuum ultraviolet (VUV) photon energies were used in laser desorption postionization mass spectrometry (LDPI-MS) to analyze biofilms comprised of binary cultures of interacting microorganisms. The effect of photon energy was examined using both tunable synchrotron and laser sources of VUV radiation. Principal components analysis (PCA) was applied to the MS data to differentiate species in Escherichia coli-Saccharomyces cerevisiae coculture biofilms. PCA of LDPI-MS also differentiated individual E. coli strains in a biofilm comprised of two interacting gene deletion strains, even though these strains differed from the wild type K-12 strain by no more than four gene deletions each out of approximately 2000 genes. PCA treatment of 7.87 eV LDPI-MS data separated the E. coli strains into three distinct groups, two “pure” groups, and a mixed region. Furthermore, the “pure” regions of the E. coli cocultures showed greater variance by PCA at 7.87 eV photon energies compared to 10.5 eV radiation. This is consistent with the expectation that the 7.87 eV photoionization selects a subset of low ionization energy analytes while 10.5 eV is more inclusive, detecting a wider range of analytes. These two VUV photon energies therefore give different spreads via PCA and their respective use in LDPI-MS constitute an additional experimental parameter to differentiate strains and species.
I. Introduction
Many microorganisms live within complex surface associated communities known as biofilms.1 Biofilms can be comprised of monocultures or more commonly, exist as consortia of multiple interacting species.2, 3 Consortial biofilms can exhibit complex interspecies relationships and dependencies that are subject of many ongoing investigations.4, 5 Controlling problematic medical or beneficial environmental multispecies biofilms is challenging due to limited knowledge of the critical interactions occurring between the microorganisms organized within microscale structures. Spatially differentiating strains or species in a mixed culture biofilm can provide useful information, such as localization of available metabolic potential as well as provide insight into the competitive nature of the metabolic interactions.6, 7
Escherichia coli and Saccharomyces cerevisiae are arguably the best-studied prokaryotic and eukaryotic microorganisms, respectively, and serve as ideal model systems for developing techniques to examine biofilm interactions. The current study employs two distinct binary cocultures designed to mimic a naturally occurring producer-consumer ecological motif which revolves around exchanges of metabolites between microorganisms.5 Both cocultures have a glucose-oxidizing producer member, either an E. coli deletion mutant8 or the baker’s yeast S. cerevisiae, which cross feed metabolic byproducts such as acetate or ethanol to a glucose-negative E. coli consumer strain which acts as the system’s scavenger.
Multivariate analysis can facilitate the processing of large data sets from the chemical analysis of biofilms and other intact biological samples. Principal component analysis (PCA) is the most commonly used and widely reported of the various multivariate analysis methods. PCA decomposes data with correlated measurements into a new set of uncorrelated (i.e., orthogonal) variables called principal components.9 PCA is ideal for interrogating large data sets such as mass spectra,10–12 because it reduces variable dimensionality with a minimal loss of information. For these reasons, PCA is often used for classification and/or grouping of experimental results to extract useful information. Several studies have differentiated strains of microorganisms by PCA of secondary ion mass spectra (SIMS)12, 13 and matrix assisted laser desorption ionization mass spectra (MALDI-MS).14, 15
SIMS and MALDI-MS have been widely used for MS imaging, both with and without PCA, of biofilms and other intact biological samples.16–20 SIMS has the advantage of high spatial resolution. However, molecular information for many species is often lost to fragmentation in SIMS, which readily detects low mass (m/z <300) and atomic ions. MALDI-MS is less chemically destructive than SIMS and is often used to detect molecular ions of species up to several kDa mass. However, MALDI-MS has lower spatial resolution than SIMS and requires extensive sample preparation such as the addition of matrix, which can obscure species in the low mass range. Both SIMS and MALDI-MS typically require thin, electrically conductive samples. Finally, ion suppression and local fluctuations in ionization efficiency can complicate quantification in these and other methods in MS imaging.20
Some of the limitations of SIMS and MALDI-MS for the analysis of biofilms and other biological samples can be overcome using laser desorption postionization mass spectrometry (LDPI-MS), which uses vacuum ultraviolet (VUV) radiation to induce a relatively ‘soft’ single photon ionization of laser desorbed neutrals.21, 22 LDPI-MS, also referred to as two laser mass spectrometry (L2MS),23 does not require the addition of a matrix compound to enhance desorption. LDPI-MS is not sensitive to ion suppression since it detects desorbed neutrals and shows some advantages for quantification.24 Furthermore, LDPI-MS can readily analyze thick, electrically insulating samples.25
The ability of LDPI-MS to detect intact parent ions of molecular species depends upon the extent of energy transfer during the separate laser desorption and single photon ionization steps. Prior LDPI-MS work showed that the higher VUV photon energies in the range of 7.8 – 12.5 eV improved sensitivity and produce intense molecular ion signal, but leads to formation of fragment ions and background gas ions.26, 27 Because the 10.5 eV photon energy appeared to provide the optimal balance between improved sensitivity and minimal fragmentation, a more robust laboratory source of 10.5 eV radiation was recently developed.25
The present study utilized 7.87 to 10.5 eV VUV photon energies from both tunable synchrotron and laser sources for LDPI-MS to analyze biofilms comprised of binary cultures of interacting microorganisms. PCA was applied to the MS data to differentiate species in E. coli-S. cerevisiae biofilms and to differentiate individual E. coli strains in a biofilm comprised of two interacting gene deletion strains. Clear spatial separation of both species as well as both E. coli strains, based on their distinct metabolic states, was demonstrated by PCA of LDPI-MS of intact biofilms.
II. Experimental Details
A. Biofilm growth and sample preparation
Biofilm culturing conditions were identical to those reported previously.25 The two E. coli K-12 deletion mutant strains (403G100-ΔptsGΔptsMΔglkΔgcd expressing the reporter protein ds-tomato and 307G100-ΔaceAΔldhAΔfrdA expressing reporter protein citrine, are termed here the tomato and citrine strain, respectively) and their growth conditions were described previously.8 The yeast, S. cerevisiae, was grown using a similar protocol. The term monoculture refers to a biofilm sample with only one species or strain. The term coculture refers to a binary culture of two genetically distinct microorganisms that have been grown together on the same substrate. The coculture biofilms grown here were inoculated initially a few mm apart by two monocultures, then allowed to grow towards each other until they visually overlapped. LDPI-MS analysis of coculture samples was performed on three distinct regions: two spots comprised predominantly of one type of microbe (“pure” region at the outer edges of the coculture sample) and at the center of the sample where the two species visually overlapped (“mixed” region). All biofilms used for the study were grown for an identical time period of 96 hours. All microbes were in stationary phase. Monoculture and coculture biofilms were grown under identical experimental conditions, to eliminate any variation due to different growth phases.
Biofilms were grown on insulating polycarbonate membranes, the membranes were then adhered to a stainless steel plate with copper tape for introduction into vacuum for MS analysis. Biofilms were also blotted onto stainless steel plates: the blots were then introduced into vacuum.
B. LDPI-MS instrumentation
LDPI-MS analysis was carried out on customized instruments utilizing either quasi-continuous synchrotron or pulsed laser VUV postionization sources. Details of the synchrotron LDPI-MS instrument have been reported previously.26–29 In brief, the synchrotron LDPI-MS was located on the Chemical Dynamics Beamline at the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA)30 and consisted of a commercial SIMS instrument (TOF.SIMS 5, ION-TOF Inc., Munster, Germany) modified by the addition of a 349 nm Nd:YLF pulsed desorption laser (Spectra-Physics Explorer, Newport Corporation, Irvine, CA). This laser was operated at 2500 Hz repetition rate with a spot size of ~30 µm diameter and laser desorption peak power density of 1 to 10 MW/cm2. Tunable VUV synchrotron radiation in the range of 7.87 – 10.5 eV photon energy was additionally introduced into this instrument for single photon ionization of laser desorbed neutrals.
The laser LDPI-MS at the University of Illinois at Chicago was also described previously.22, 25 A 349 nm Nd:YLF laser (Spectra-Physics Explorer) was used for desorption at 10 or 100 Hz repetition rate which depended on the VUV ionization laser. The desorption laser beam was used at peak power density of ~300 MW/cm2 with a ~50 µm diameter beam on the biofilm. Laser postionization was carried out at two fixed photon energies: 7.87 and 10.5 eV. A 157.6 nm fluorine laser (Optex Pro, Lambda Physik, Ft Lauderdale, FL) operating at 100 Hz was used for 7.87 eV postionization with a cross sectional area of 2 × 1 mm2 and an energy of ~100 µJ/pulse. 10.5 eV laser postionization was performed with a 118 nm beam generated by tripling the third harmonic of a Nd:YAG laser (355 nm, ~20 mJ, 5 ns, Tempest, New Wave Research, Fremont, CA) in a Xe gas cell at 6.5 Torr pressure, as described previously.25 The resulting photoions were pulse extracted and analyzed using a 340 ppm mass accuracy reflectron TOF which is similar to that described previously.25 The data acquisition software was partially described elsewhere with respect to its use on a different instrument.31
The sample stage on the synchrotron LDPI-MS was rastered to analyze a ~3 mm2 area for each biofilm sample with 10 laser shots per spot before moving to an fresh spot for repeated analysis such that ~1.2 × 105 laser shots were used for collecting each displayed synchrotron mass spectrum. The laser LDPI-MS utilized a sample stage that was continuously moving at a speed of 0.05 mm/s while the desorption laser scanned over the sample for a total area of analysis of 1 mm2. Mass spectra were averaged over the entire analyzed area such that ~4 × 104 laser shots were used to collect each displayed laser LDPI mass spectrum.
C. Data acquisition and analysis
A custom program was designed in-house for converting full mass spectra to integer m/z peak values similar to those used for electron impact MS database searching.32 No peak selections were made. Rather, the entire raw mass spectrum was aligned to integer m/z values, then zeroes were inserted as intensity values for peaks where no ion signal was observed above the assigned signal threshold of ~35 mV. Ion intensities were normalized prior to PCA by setting the most intense peak to unity, thereby minimizing potential fluctuations due to instrumental factors such as modulating detector efficiencies. Finally, ion intensities at each integer m/z 50–700 value for each sample were exported to a spreadsheet. A nonlinear iterative partial least squares algorithm (“princomp” function, MATLAB, The MathWorks Inc., Natick, MA) was used to perform PCA33, 34 on the n × p data matrix where n represents the number of experiment trials for each strain and p represents the ion intensities written to the aforementioned spreadsheet for each integer m/z value. PCA was performed after mean centering of the columns of the data matrix. Each data set used in the analysis was replicated at least three times and a minimum of 15 spectra of each microbe type was used to compose the original data matrix.
III. Results
A. Effect of VUV Photon Energy on Analysis of Monoculture Biofilms
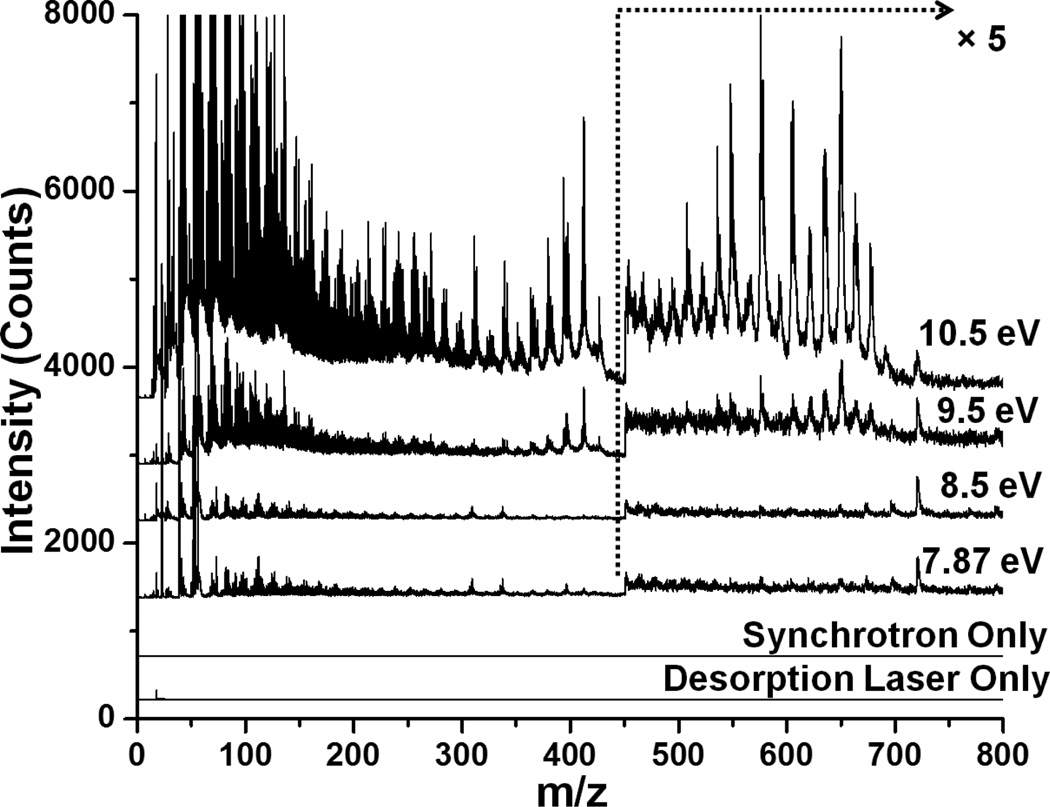
Blots of yeast monoculture biofilms were analyzed by synchrotron LDPI-MS using 7.87, 8.5, 9.5 and 10.5 eV VUV photon energies with the results shown in Figure 1. The synchrotron LDPI-MS of yeast monocultures showed that increasing photon energy resulted in more peaks in the mass spectra and higher overall signal intensity. 10.5 eV showed more peaks for molecular species as well as more fragments compared to the lower photon energies. However, fragmentation increased significantly at photon energies between 10.5 and 15 eV (data not shown). Overall, 10.5 eV photon energy displayed an optimal balance between sensitivity and fragmentation, in agreement with prior experiments.26, 27, 35 Similar conclusions were reached from 7.87 – 10.5 eV synchrotron LDPI-MS of E. coli (tomato strain) monoculture blots (full mass spectra shown in Supplementary Information). The mass spectral peaks observed for E. coli and yeast monocultures were tabulated with the VUV photon energies at which they were first observed by synchrotron LDPI-MS (see Supplementary Information).
Figure 1.
7.87 – 10.5 eV synchrotron laser desorption postionization mass spectra (LDPI-MS) of blotted yeast monoculture biofilms. The bottom two traces show the controls performed with only synchrotron photoionization or desorption laser.
Prior synchrotron LDPI-MS studies26, 27 led to the recent development of a refined laboratory VUV photoionization source that operates at 10.5 eV25 to complement the 7.87 eV fluorine excimer laser source.22 These two laboratory laser VUV sources were used to collect 7.87 and 10.5 eV laser LDPI-MS data from yeast and E. coli monocultures. 10.5 eV laser LDPI-MS data for blotted monoculture biofilms (yeast or E. coli) was collected and compared with the similar spectra collected by 10.5 eV synchrotron LDPI-MS (see Supplementary Information). The synchrotron LDPI-MS data of the blotted samples showed overall higher S/N compared to that from laser LDPI-MS.
The data traces labeled “Desorption Laser Only” and “Synchrotron Only” in Figure 1 demonstrate the absence of any background signal arising from direct ionization via the desorption laser or VUV postionization of background or sublimed gaseous neutrals, respectively. Prior control experiments with the 10.5 eV laser LDPI-MS established the presence of single photon ionization and ruled out both direct ions from laser desorption and photoelectron ionization effects due to the residual 355 nm beam used to generate VUV radiation.25 Photoelectron ionization was previously ruled out in the synchrotron LDPI-MS.27 The similarity of the spectra of 7.87 eV synchrotron and laser LDPI-MS ruled out photoelectron ionization in the latter case.
B. Analysis of intact biofilms vs. blotted samples
It is technically convenient to grow biofilms on polymer membranes, but these insulating membranes can complicate analysis by MS imaging strategies. Membrane biofilm analyses were attempted on the synchrotron LDPI-MS instrument, but reproducible MS signal was elusive due to what appeared to be excessive sample charging of these electrically insulating membranes. However, intact membrane biofilms could be analyzed directly in vacuum by laser LDPI-MS, as discussed below.
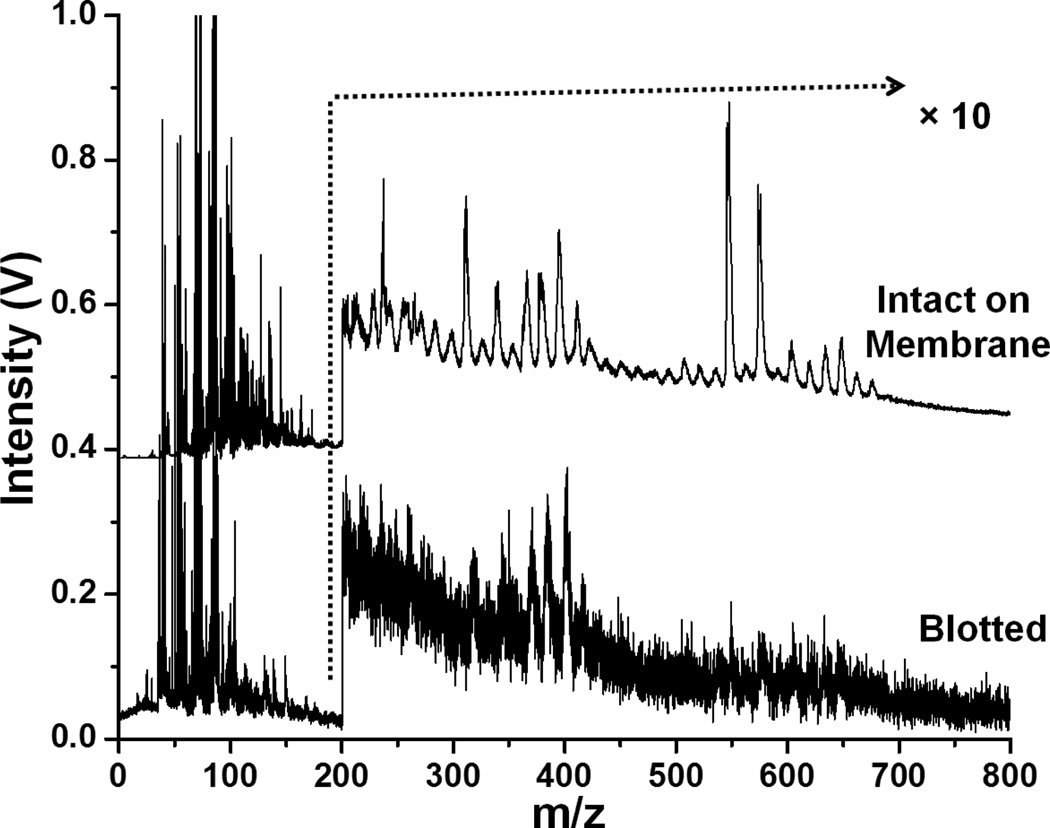
Figure 2 shows 10.5 eV laser LDPI mass spectra of blotted and intact biofilm samples from yeast monocultures. There was ~8× higher S/N for the intact membrane samples compared to the blotted biofilms. Although the same peaks were observed for both samples, overall peak intensity was higher for membrane samples. Additionally, peaks at m/z 600 – 700 were not discernible in the blotted sample due to low S/N.
Figure 2.
10.5 eV laser LDPI-MS of yeast monoculture biofilms using two different sample preparation techniques. Spectra are normalized.
Direct comparison was also made for membrane biofilms analyzed with laser LDPI-MS vs. blotted biofilms analyzed with synchrotron LDPI-MS: the different spectra showed similar peaks, albeit with differences in S/N and absolute signal intensity. The synchrotron MS data showed more peaks in the low mass (< m/z 300) region.
Intact membrane biofilms were not observed to delaminate in vacuum and maintained their structural integrity. However, the blotted biofilms occasionally suffered from delamination in vacuum which adversely impacted the synchrotron LDPI-MS. The continuous synchrotron radiation readily ablated delaminating pieces since the VUV beam passed very close to the top of the sample, leading to signal spikes near m/z 270 in the MS (see Supplementary Information). Ablation signal was not observed in laser LDPI-MS since the pulsed VUV beam passed farther above the sample, missing the delaminated pieces of biofilm. In any case, ablation would occur to a lesser extent by the pulsed VUV laser beam whose duty cycle was 10−6 at 7.87 eV and 10−7 at 10.5 eV (vs. near unity for the quasi-continuous synchrotron radiation).
C. Coculture multispecies biofilms
10.5 eV synchrotron LDPI-MS were recorded at three distinct regions of blotted E. coli and yeast coculture biofilms (see Supplementary Information). The “pure” E. coli and yeast regions of the biofilms generated spectra that generally appeared similar to those from the corresponding monocultures. However, synchrotron LDPI-MS of the mixed region of the coculture biofilms tended to vary from one sample to the next. This variability might have resulted from the limits of sample preparation: the clear boundaries of the microbial species were not visually apparent in the blotted samples, making it difficult to assign a given region to a specific species. Additionally, synchrotron LDPI-MS were acquired with desorption laser scanning over a relatively large, ~3 mm2 area for each region, increasing the possibility of overlap of the different regions.
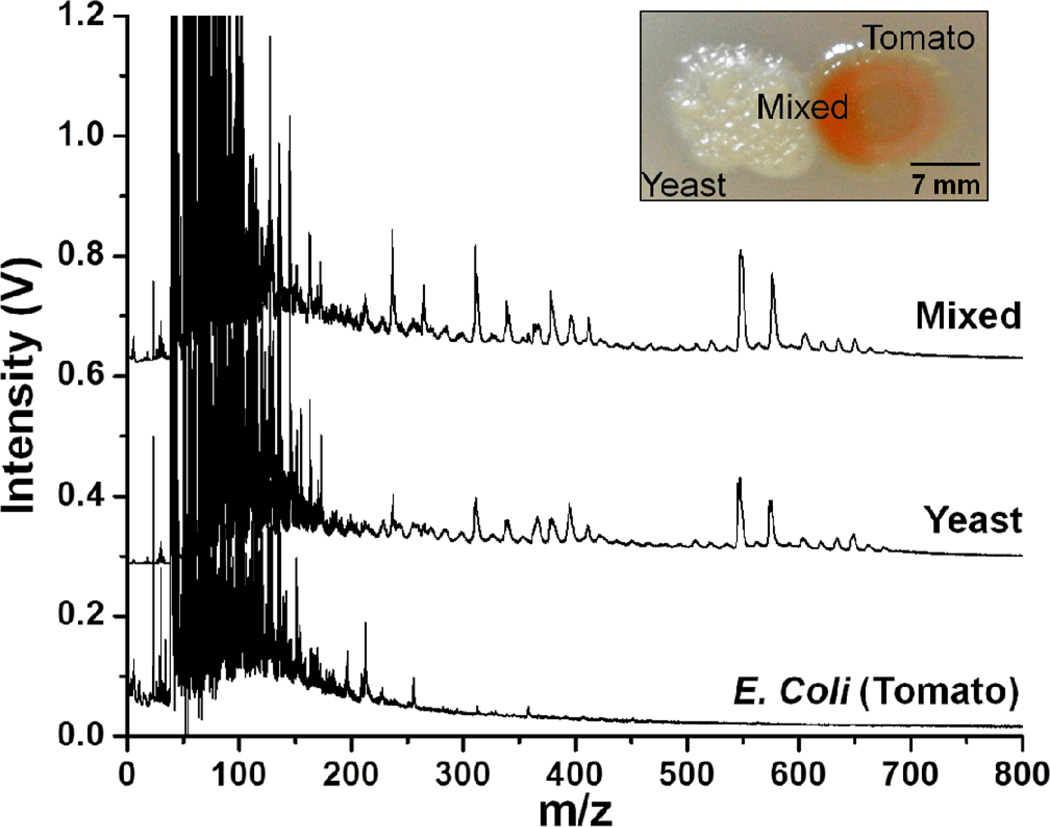
By contrast, the laser LDPI-MS allowed analysis of a ~1 mm2 area of an intact membrane biofilm with relatively high S/N. Therefore, coculture E. coli and yeast biofilms were studied using both the 7.87 and 10.5 eV laser photoionization sources, with Figure 3 showing the latter result. The trace labeled “Mixed” in Figure 3 refers to the region where the two species overlap. The “pure” regions of E. coli and yeast could be readily distinguished by visual examination of the intact membrane biofilms and the mass spectra at both photon energies appeared similar to those of their respective monocultures. The mass spectra for the mixed region showed peaks from both of the species. 10.5 eV laser LDPI-MS showed more peaks in the high mass region (m/z > 400) compared to 7.87 eV data, indicating more molecular species were ionized at the higher photon energy (see Supplementary Information).
Figure 3.
10.5 eV laser LDPI-MS of coculture E. coli (tomato strain) and yeast multispecies membrane biofilm. “Mixed” indicates the region of overlap between the two species while other spectra correspond to “pure” regions of biofilms, as labeled in the inset photo of a typical coculture biofilm.
D. PCA of LDPI-MS for strain and species differentiation
The results above demonstrated the capability of LDPI-MS to study monoculture biofilms as well as multispecies coculture biofilms (see also Supplementary Information). The mass spectral data however, did not show much difference between the analyzed microbes. Additionally, the low mass region (m/z < 250) which is of great interest for metabolite study was too crowded to visually observe any obvious differences among strains. PCA was therefore used as a statistical tool to reduce the data size without significant information loss and for better visualization of the data. The approach was optimized by first applying PCA to the clearly distinct MS data of E. coli and yeast in a coculture biofilm samples (see Supplementary Information). Given that these two microbes showed obvious mass spectral differences for the “pure” regions of each species, it is not surprising that applying PCA to the data also readily distinguished the two species.
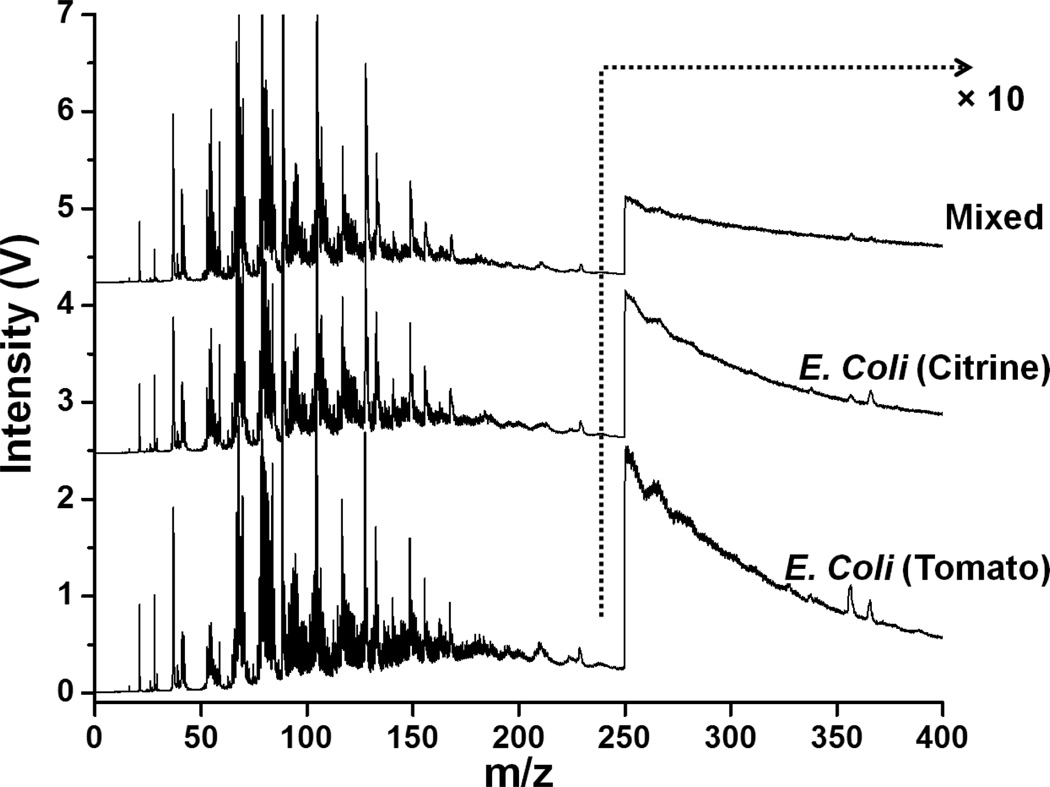
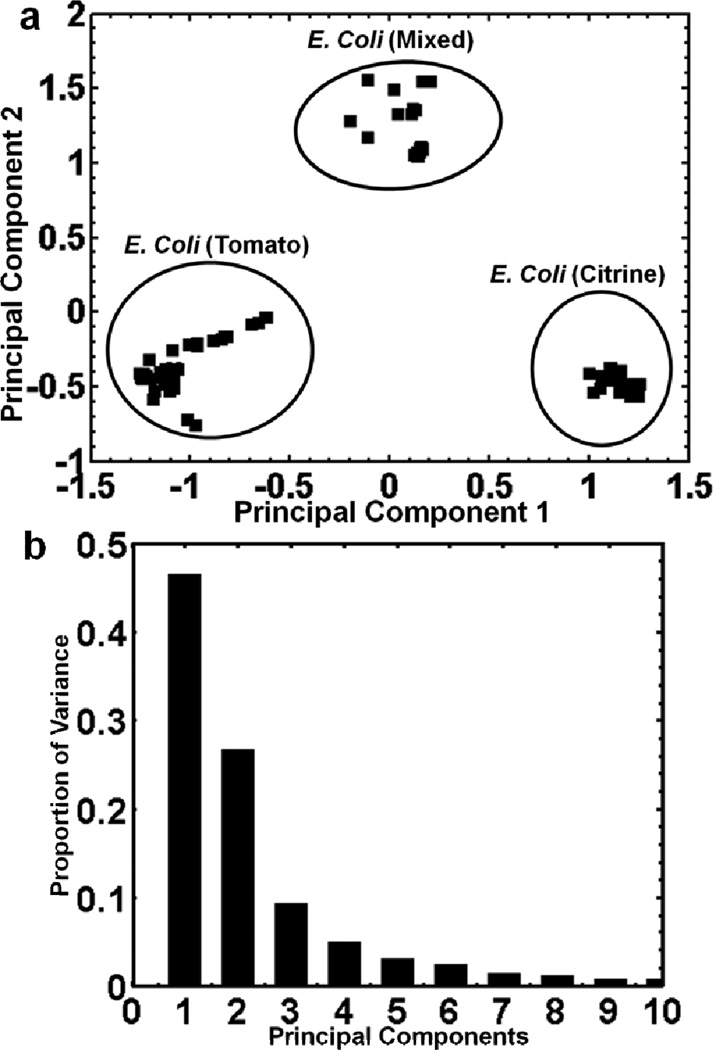
The optimized PCA protocol was then applied to a more challenging system wherein a multistrain E. coli membrane biofilm was examined. Both 7.87 eV and 10.5 eV laser LDPI-MS data of the genetically similar tomato and citrine E. coli strains were collected, with the former shown in Figure 4 and the latter reported previously.25 Figures 5a shows a plot of principal component 1 vs. principal component 2 for the 7.87 eV laser LDPI-MS data. While the mass spectra of the two strains in coculture samples showed only minor visual differences in the peak pattern, PCA treatment of the “pure” regions of the sample resulted in grouping the two strains separately. Furthermore, the mixed region was clearly distinguished from the two “pure” regions suggesting metabolic interactions resulted in altered physiologies. Figure 5b is a scree plot9 which shows the variance of the entire data set with respect to the principal components. The scree plot indicates that ~75% of the variance for the data is represented by its first two principal components. In addition, hierarchical cluster analysis was performed to the aforementioned 7.87 eV mass spectral data (see Supplementary Information) that showed similar clustering of the strains to that obtained by PCA. Similarly, PCA was performed on the corresponding 10.5 eV MS data (see Supplementary Information): it also distinguished the two “pure” regions of the biofilm, with most variance along the first principal component.
Figure 4.
7.87 eV laser LDPI-MS of coculture E. coli (tomato and citrine) multistrain membrane biofilm. “Mixed” indicates the region of overlap between the two strains.
Figure 5.
a) Principal component analysis of 7.87 eV laser LDPI-MS of the three different regions of a coculture multistrain E. coli (tomato and citrine strains) membrane biofilm. b) Scree plot showing variance of the entire data set with respect to the principal components.
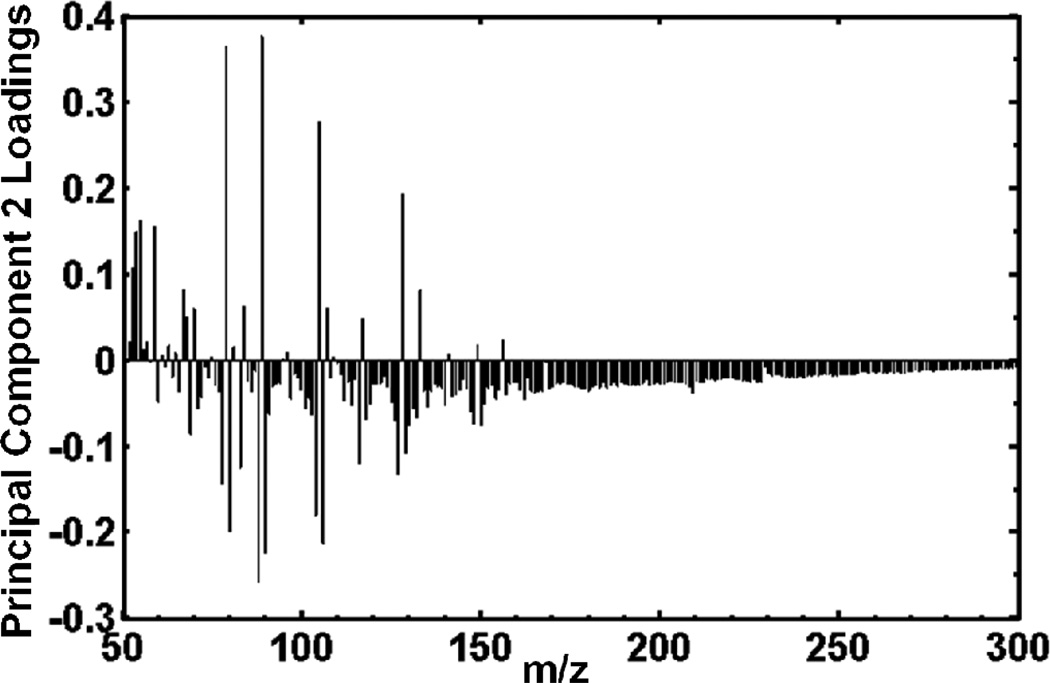
Figure 6 is the principal component 2 loadings plot, correlating which peaks in the mass spectra are most responsible for the differences seen by PCA between the mixed region and the two “pure” regions of the 7.87 eV laser LDPI-MS. In general, peaks with positive loadings along a given PC axis will show a higher relative intensity in samples with positive scores along the same PC axis (and negative loadings are similarly correlated with negative scores).12 Figure 5a shows that the E. coli mixed region has a positive score while the two pure regions have negative scores along principal component 2. Therefore, positive peaks in Figure 6 loadings plot for PC 2 correspond with the peaks that are more abundant in the mixed region (Figure 5a). Figure 6 and the biplot (not shown) indicate that the mass spectral peaks that contributed the most to the separation of the mixed region from the two pure regions were m/z 54, 55, 59, 60, 67, 79, 84, 89, 105, 128 and 133. Similar analysis of the 7.87 eV data for the two pure regions indicated that peaks at m/z 55, 56, 67, 71, 78, 80, 88, 90, 104, 106, 127 and 129 contributed to the separation of the two strains along the first principal component. PCA of 10.5 eV laser LDPI-MS data indicated that peaks at m/z 53, 65, 75, 77, 79, 81, 92, 94, 108, 216, 258 and 285 contributed to the separation of the two strains. However, it cannot be determined in these experiments which of these peaks are due to parent vs. fragment ions.
Figure 6.
Principal component 2 loadings plot for the entire 7.87 eV laser LDPI-MS data set of the coculture multistrain E. coli biofilm.
PCA was also applied to the synchrotron LDPI-MS data, but no coherent results could be obtained from the analysis. One limitation of the synchrotron LDPI-MS data analysis was simply that too few spectra had been collected for a useful data matrix, a problem to be overcome by further experiments.
IV. Discussion
A. Distinguishing species, strains, and metabolic states by PCA of LDPI-MS data
Different species as well as separate strains and interfacial regions of coculture biofilms were distinguished by LDPI-MS detection of endogenous species followed by discrimination of the data by PCA. Additionally, hierarchical cluster analysis of the data showed that the strains and metabolic states of coculture biofilms clustered separately, with the results corresponding with those for PCA (see Supplementary Information). This is especially remarkable for the multistrain system because the strains differed from the wild type K-12 strain by no more than four gene deletions each out of approximately 2000 genes8 and visual examination of the mass spectral data showed barely any differences.25 PCA treatment of 7.87 eV LDPI-MS data separated the strains into three distinct groups (see Figure 5a): two “pure” groups and a mixed region. Furthermore, the “pure” regions of the cocultures showed greater variance by PCA when analyzed by 7.87 eV photon energies than by 10.5 eV radiation. It is noteworthy that the E. coli mixed region (Figure 5a) had a positive score along principal component 2 which resulted in a grouping outside of the pure tomato and citrine regions.
These results indicate that the mixed region of the multistrain system constituted a different collective metabolic profile than a simple summation of the pure metabolisms, possibly due to inter-strain interactions via metabolite or quorum sensing-like exchange. Use of multiple trials supports the claim of a distinct metabolic state in the mixed region that is not due to contaminants or other matrix effects. In particular, salt gradients are not expected within the multistrain biofilms. Furthermore, salt effects are minimized by the detection of desorbed neutrals in LDPI-MS. LDPI-MS was also able to distinguish the bacteria from the yeast studied here both without and with the application of PCA, both significant accomplishments. These results establish LDPI-MS as another MS tool available to probe local metabolic states and metabolite exchange in microbial systems.6
The principal component 2 loadings plot for the 7.87 eV laser LDPI-MS data (Figure 6) revealed several mass spectral that contributed the most to the separation of the strains. Exact mass measurements and or tandem MS are required to conclusively assign peaks to any compound, so such assignments are generally not attempted here (see Supplementary Information). Nevertheless, hypotheses are made regarding a few of the peaks contributing to the separation of the strains (Figure 5 and 6). For example, the m/z 60 peak showed a higher relative intensity in the mixed region (Figure 6) and contributed to the separation of the mixed region from the “pure” regions. This peak might have arisen from the molecular ion of acetate which is a common E. coli metabolic byproduct of glucose metabolism which can be cross-fed to the tomato scavenger strain as a substrate.8 The principal component 1 loadings plot of the 10.5 eV laser LDPI-MS showed that m/z 285 contributed to the difference between the two E. coli strains. M/z 285 could be attributed to the stearic acid parent ion, which is a common constituent of the E. coli cell walls and membranes.25 It is proposed that the two strains’ metabolisms create locally different chemical microenvironments that influence biofilm composition.
Both the synchrotron and laser LDPI-MS indicated that different classes of molecules can be targeted with different photon energies. New peaks appeared with increasing photon energy (see Supplementary Information), as expected since many organic compounds have ionization energies in the 9 – 10 eV range and single photon ionization requires VUV photon energies in excess of a molecule’s ionization energy.22, 36 Comparison of the 7.87 and 10.5 eV data is consistent with the expectation that the lower photon energy selects a subset of low ionization energy analytes while 10.5 eV is more inclusive, detecting across a wider range of analytes. These two VUV photon energies therefore give different spreads via PCA and they constitute an additional experimental parameter to differentiate strains and species. Many metabolites which participate in crossfeeding or quorum sensing between interacting strains and species have relatively low molecular weights, so their molecular ions will be found in the low mass region. The lower mass region is of particular note in the 7.87 eV LDPI-MS of the E. coli cocultures.
B. Comparison of sample preparation methods
Thick, intact biofilm samples grown on insulating membranes were successfully analyzed using laser LDPI-MS. Polycarbonate membranes provided a surface for robust growth of coculture biofilms and can produce visually obvious boundaries between different species or strains. However, biofilms grown on membranes were ~0.5 – 1 mm thick and displayed topographical features that can hinder analysis by SIMS and MALDI-MS imaging.
Prior LDPI-MS studies examined biofilms grown using drip flow reactors,26 but coculture biofilms grown by drip flow tend to lack clear boundaries between segregated species making analysis of pure cultures impossible without additional parallel experiments. Significantly higher laser desorption energy was required to analyze intact membrane biofilms compared to the blotted biofilms. This might have resulted from the thickness of the membrane biofilms and the effective absence of an immediately adjacent metal substrate which otherwise can undergo rapid heating to assist desorption.29, 37
Synchrotron LDPI-MS was able to successfully analyze blots of biofilms, which can also be analyzed by other MS imaging methods,16 but could not analyze intact membrane biofilms. Laser LDPI-MS of intact membrane biofilms showed overall higher signal intensity than from blotted samples. This could be due to the difference in desorption efficiency for the two samples. By contrast, blotting of the biofilm resulted in loss of structural integrity and reduced sample volume that decreased the overall signal intensity. Furthermore, the efficiency of chemical transfer during blotting, including differential transfer, cannot be ruled out as a contributing factor.
C. Comparison of data analysis and instrumental methods
Several studies have been reported which applied PCA to MALDI-MS data to obtain differentiation of species or strains as well as biomarker profiling.14, 15 However, it is noteworthy that differences between MALDI mass spectra due to ion suppression, matrix peak intensities, and fluctuating baseline might lead to unwarranted sample separation during PCA. By contrast, LDPI-MS can analyze the low mass region of coculture biofilms free of ion suppression, without addition of any interfering matrix, and without any background subtraction.
SIMS has the advantage of high spatial resolution and high sensitivity in the low mass region, both useful for studying metabolites and other small endogenous species. However, MS data for masses over m/z 300 can be difficult to observe in SIMS, limiting the application of SIMS to a reduced set of target molecules.12, 13, 18, 38 The results presented here showed some LDPI-MS peaks up to m/z 1000, indicating its potential use for target species with a larger mass range than commonly available to SIMS. Furthermore, LDPI-MS should induce less fragmentation in molecular species than does SIMS, so LDPI-MS data should better represent the composition of the lower mass region. Nevertheless, the relatively low abundance of LDPI-MS peaks at m/z >300 is attributed larger to fragmentation from energy transfer during laser desorption rather than single photon ionization.39, 40 Relatively low species abundance and difficulty in desorbing some of the neutral species from intact biofilm samples also likely contributed to the absence of higher mass peaks. A recent study described how LDPI-MS provides complementary chemical information to SIMS for the analysis of soil organic matter.40
Atmospheric pressure-based methods such as nano-desorption electrospray ionization (nano-DESI)41, 42 and laser ablation electrospray ionization (LAESI)19 allow for MS imaging of biofilms without sample deformation or the loss of volatile species that is possible in vacuum. However, such electrospray ionization-based methods suffer from the same effects of ion suppression, ion-molecule reactions, and local fluctuations in ionization efficiency that can complicate identification and quantification of species in SIMS.20 LDPI-MS seeks to overcome the shortcomings of direct ion formation by postionization of neutrals at pressures low enough to avoid subsequent ion-molecule reactions, albeit with the potential loss of volatile species prior to ionization. Unlike methods that detect direct ions, salt effects are minimized by the detection of desorbed neutrals in LDPI-MS.
With its ability to analyze intact biological samples with reasonable spatial resolution, LDPI-MS can be used to study a range of biological samples such as infected medical implants, dental composites, or tooth samples. Spatial resolution here was limited to the mm-scale due to the relatively low signal that required collection of spectra from relatively large areas of the biofilms. Prior work reported a spatial resolution of ~50 µm is achievable with the laser LDPI-MS instrument as limited by mechanical vibrations and the low repetition rate of the Nd:YAG laser used to pump the 10.5 eV source.25 Synchrotron LDPI-MS has demonstrated a resolution of better than 5 µm on an organic photoresist structure.29 LDPI-MS with femtosecond desorption lasers should permit micron scale resolution as well as depth profiling of biological samples,43–45 the latter a strategy only definitively demonstrated on SIMS.20 By comparison, SIMS displays submicron spatial resolution while atmospheric pressure-based methods typically allow spatial resolution in the tens of microns.18, 20
The data presented here was acquired with two different LDPI-MS instruments. The synchrotron LDPI-MS instrument used tunable VUV for postionization that allowed exploration of the effect of increasing photon energies on mass spectral profiles. The synchrotron LDPI-MS showed overall higher signal intensity and with appropriate sample preparation should allow for high resolution imaging of microbiological systems. Prior work on non-biofilm samples demonstrated the utility of tuning across a wide and continuous range of photon energies.29, 36 The 7.87 and 10.5 eV laser LDPI-MS allowed for in house data analysis and optimization as well as examination of electrically insulating samples such as membrane biofilms. Furthermore, the 7.87 eV fluorine laser and the 10.5 eV laser-based sources are both sufficiently robust that either could be implemented on commercial instrumentation such as vacuum source-based MALDI-MS instruments, converting them into LDPI-MS instruments.
The effect of varying delay time between the desorption and ionization processes was not explored in this study. However, prior work showed that sensitivity and resolution can be optimized in LDPI-MS at different mass ranges by varying delay times and other ion optical parameters, with more high mass peaks detected at longer delay times. A comparison of MS methods noted a lack of any single desorption/ionization technique that permits characterization of all components in crude oil samples.46 A similar statement can be made regarding the analysis of biofilms, which combined with the data provided above supports the argument that LDPI-MS can make a valuable contribution to the analysis of these and other intact biological samples.
Supplementary Material
Acknowledgements
The authors acknowledge the assistance of Anjan Roy in developing the multivariate analysis and thank Gulsah Uygur for useful discussions. This work was supported by the National Institute of Biomedical Imaging and Bioengineering via grant EB006532. MA, TH, SL, and the Advanced Light Source were supported by the Director, Office of Energy Research, Office of Basic Energy Sciences, Chemical Sciences Division of the U.S. Department of Energy under contract No. DE-AC02-05CH11231. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering, the National Institutes of Health, or the Department of Energy.
Footnotes
Supplementary Information
The following figures and associated text are provided in the Supplementary Information.
Figure S1: 7.87 to 10.5 eV synchrotron LDPI-MS of E. coli (tomato) strain blotted monoculture biofilms.
Figure S2: 10.5 eV laser and synchrotron LDPI-MS of blotted monoculture biofilms using two different instruments.
Table S1: List of mass peaks (m/z) first observed at 7.87, 9.5, and 10.5 eV photon energies by synchrotron LDPI-MS of blots of E. coli bacteria and S. cerevisiae yeast monocultures
Figure S3: 10.5 eV synchrotron LDPI-MS of different regions of a blotted coculture biofilm.
Figure S4: 7.87 eV laser LDPI-MS of three regions of E. coli (tomato strain) and yeast coculture membrane biofilm.
Figure S5: Principal component 1 loadings plot for the 7.87 eV laser LDPI-MS data set of the coculture multistrain biofilm.
Figure S6: Principal component analysis of 10.5 eV laser LDPI-MS of a E. coli (tomato strain) and yeast coculture membrane biofilm.
Figure S7: a) Principal component analysis of 10.5 eV laser LDPI-MS of a coculture tomato and citrine E. coli membrane biofilm. b) Scree plot showing the variance of the data with respect to the principal components.
Figure S8: PCA PC1 loadings plot from 10.5 eV laser LDPI-MS data for coculture E. coli biofilm analyzed in Figure S6.
Figure S9: Hierarchical cluster analysis (HCA) for 7.87 eV LDPI-MS data.
References
- 1.Bandara HMHN, Yau JYY, Watt RM, Jin LJ, Samaranayake LP. J. Med. Microbiol. 2009;58:1623–1631. doi: 10.1099/jmm.0.012989-0. [DOI] [PubMed] [Google Scholar]
- 2.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. Curr. Biol. 2008;18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner K, You L, Arnold FH. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Carlsson J. Adv. Dental Res. 1997;11:75–80. doi: 10.1177/08959374970110012001. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein H, Carlson RP. Comput. Struct. Biotechnol. J. 2012;3:e201210017. doi: 10.5936/csbj.201210017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phelan VV, Liu WT, Pogliano K, Dorrestein PC. Nat. Chem. Biol. 2012;8:26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Momeni B, Brileya KA, Fields MW, Shou W. eLife Sciences. 2013;2 doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein HC, Paulson SD, Carlson RP. J. Biotechnol. 2012;157:159–166. doi: 10.1016/j.jbiotec.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otto M. Chemometrics: Statistics and computer application in analytical chemistry. New York: Wiley-VCH; 2007. [Google Scholar]
- 10.Tyler BJ, Rayal G, Castner DG. Biomaterials. 2007;28:2412–2423. doi: 10.1016/j.biomaterials.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shrestha B, Patt JM, Vertes A. Anal. Chem. 2011;83:2947–2955. doi: 10.1021/ac102958x. [DOI] [PubMed] [Google Scholar]
- 12.Graham DJ, Castner DG. Biointerph. 2012;7:49. doi: 10.1007/s13758-012-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyler BJ, Rangaranjan S, Möller J, Beumer A, Arlinghaus HF. Appl. Surf. Sci. 2006;252:6712–6715. [Google Scholar]
- 14.Chen P, Lu Y, Harrington PB. Anal. Chem. 2008;80:1474–1481. doi: 10.1021/ac7018798. [DOI] [PubMed] [Google Scholar]
- 15.Qian J, Cutler JE, Cole RB, Cai Y. Anal. Bioanal. Chem. 2008;392:439–449. doi: 10.1007/s00216-008-2288-1. [DOI] [PubMed] [Google Scholar]
- 16.Watrous JD, Dorrestein PC. Nat. Rev. Microbiol. 2011;9:683–694. doi: 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blaze M.T M, Aydin B, Carlson RP, Hanley L. Analyst. 2012;137:5018–5025. doi: 10.1039/c2an35922g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trouillon R, Passarelli MK, Wang J, Kurczy ME, Ewing AG. Anal. Chem. 2012;85:522–542. doi: 10.1021/ac303290s. [DOI] [PubMed] [Google Scholar]
- 19.Vertes A, Hitchins V, Phillips KS. Anal. Chem. 2012;84:3858–3866. doi: 10.1021/ac2029997. [DOI] [PubMed] [Google Scholar]
- 20.Fletcher JS, Vickerman JC. Anal. Chem. 2013;85:610–639. doi: 10.1021/ac303088m. [DOI] [PubMed] [Google Scholar]
- 21.Hanley L, Zimmermann R. Anal. Chem. 2009;81:4174–4182. doi: 10.1021/ac8013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akhmetov A, Moore JF, Gasper GL, Koin PJ, Hanley L. J. Mass Spectrom. 2010;45:137–145. doi: 10.1002/jms.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabbah H, Morrow AL, Pomerantz AE, Zare RN. Ener. Fuels. 2011;25:1597. [Google Scholar]
- 24.Blaze M.T M, Akhmetov A, Aydin B, Edirisinghe PD, Uygur G, Hanley L. Anal. Chem. 2012;84:9410–9415. doi: 10.1021/ac302230e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhardwaj C, Moore JF, Cui Y, Gasper GL, Bernstein HC, Carlson RP, Hanley L. Anal. Bioanal. Chem. 2012 doi: 10.1007/s00216-012-6454-0. http://dx.doi.org/10.1007/s00216-012-6454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasper GL, Takahashi LK, Zhou J, Ahmed M, Moore JF, Hanley L. Anal. Chem. 2010;82:7472–7478. doi: 10.1021/ac101667q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaze M.T M, Takahashi LK, Zhou J, Ahmed M, Gasper GL, Pleticha FD, Hanley L. Anal. Chem. 2011;83:4962–4969. doi: 10.1021/ac200693h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi LK, Zhou J, Wilson KR, Leone SR, Ahmed M. J. Phys. Chem. A. 2009;113:4035–4044. doi: 10.1021/jp810408v. [DOI] [PubMed] [Google Scholar]
- 29.Kostko O, Takahashi LK, Ahmed M. Chem., Asian J. 2011;6:3066–3076. doi: 10.1002/asia.201100419. [DOI] [PubMed] [Google Scholar]
- 30.Heimann PA, Koike M, Hsu CW, Blank D, Yang XM, Suits AG, Lee YT, Evans M, Ng CY, Flaim C, Padmore HA. Rev. Sci. Instrum. 1997;68:1945–1951. [Google Scholar]
- 31.Cui Y, Moore JF, Milasinovic S, Liu Y, Gordon RJ, Hanley L. Rev. Sci. Instrum. 2012;83:093702. doi: 10.1063/1.4750974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein S. Anal. Chem. 2012;84:7274–7282. doi: 10.1021/ac301205z. [DOI] [PubMed] [Google Scholar]
- 33.Timmins ÉM, Howell SA, Alsberg BK, Noble WC, Goodacre R. J. Clin. Microbiol. 1998;36:367–374. doi: 10.1128/jcm.36.2.367-374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodacre R, Heald JK, Kell DB. FEMS Microbiol. Lett. 1999;176:17–24. [Google Scholar]
- 35.Gasper GL, Takahashi LK, Zhou J, Ahmed M, Moore JF, Hanley L. Nucl. Instrum. Methods Phys. Res., Sect. A. 2011;649:222–224. doi: 10.1016/j.nima.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi LK, Zhou J, Kostko O, Golan A, Leone SR, Ahmed M. J. Phys. Chem. A. 2011;115:3279–3290. doi: 10.1021/jp111437e. [DOI] [PubMed] [Google Scholar]
- 37.Hanley L, Kornienko O, Ada ET, Fuoco E, Trevor JL. J. Mass Spectrom. 1999;34:705–723. doi: 10.1002/(SICI)1096-9888(199907)34:7<705::AID-JMS845>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 38.Cillero-Pastor B, Eijkel G, Kiss A, Blanco FJ, Heeren RMA. Anal. Chem. 2012;84:8909–8916. doi: 10.1021/ac301853q. [DOI] [PubMed] [Google Scholar]
- 39.Isaacman G, Wilson KR, Chan AWH, Worton DR, Kimmel JR, Nah T, Hohaus T, Gonin M, Kroll JH, Worsnop DR, Goldstein AH. Anal. Chem. 2012;84:2335–2342. doi: 10.1021/ac2030464. [DOI] [PubMed] [Google Scholar]
- 40.Liu SY, Kleber M, Takahashi LK, Nico P, Keiluweit M, Ahmed M. Anal. Chem. 2013;85:6100–6106. doi: 10.1021/ac400976z. [DOI] [PubMed] [Google Scholar]
- 41.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Anal. Chem. 2011;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rath CM, Alexandrov T, Higginbottom SK, Song J, Milla ME, Fischbach MA, Sonnenburg JL, Dorrestein PC. Anal. Chem. 2012;84:9259–9267. doi: 10.1021/ac302039u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milasinovic S, Liu Y, Gasper GL, Zhao Y, Johnston JL, Gordon RJ, Hanley L. J. Vac. Sci. Technol., A. 2010;28:647–651. doi: 10.1116/1.3397736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milasinovic S, Liu Y, Bhardwaj C, Blaze M.T M, Gordon RJ, Hanley L. Anal. Chem. 2012;84:3945–3951. doi: 10.1021/ac300557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui Y, Bhardwaj C, Milasinovic S, Carlson RP, Gordon RJ, Hanley L. ACS Appl. Mater. Interf. 2013 doi: 10.1021/am4020633. [DOI] [PubMed] [Google Scholar]
- 46.Panda SK, Brockmann K-J, Benter T, Schrader W. Rapid Comm. Mass Spectrom. 2011;25:2317–2326. doi: 10.1002/rcm.5122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.