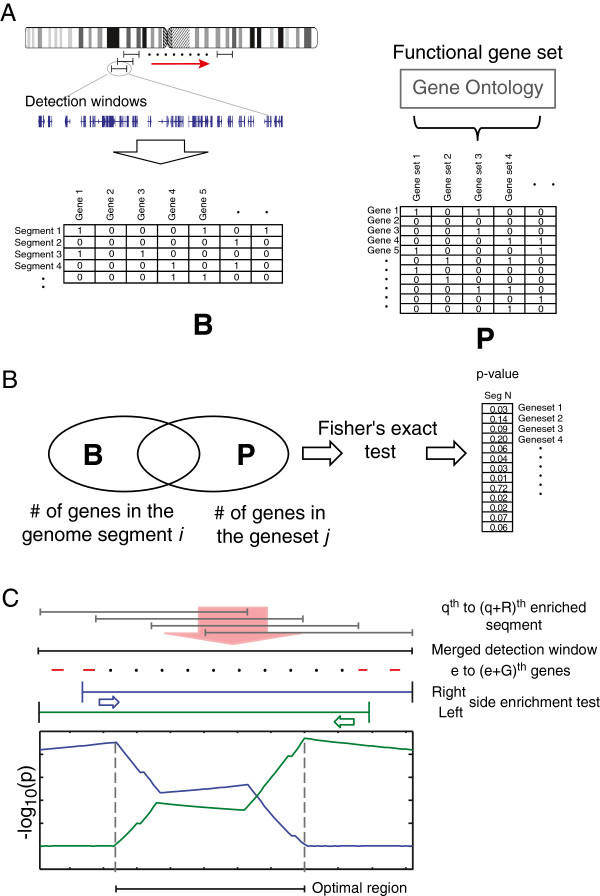
Figure 1.
Schematic diagram illustrating the enrichment analysis of spatial functional hotspots in human genome. (A) The indicator matrix B was generated by sliding the detection window along the genome. It contains the information of genes located in each segment. The matrix P records the gene sets of Gene Ontology. (B) By comparing the two matrices assessed with Fisher's exact test, the p values of gene sets in each segment were generated. The enriched functions of each segment were then identified if passed the selection criteria. (C) The nearby segments with the same enriched function were merged to a detection window. The enrichment analysis between the function and the subset of genes in the windows were performed. The subset was constructed by excluding gene by gene along the left side or right side of the genome coordinate. The position with the smallest p value of left side and right side excluding subset was defined as the boundaries of the optimal region of the functional hotspot.