Fig. 4.
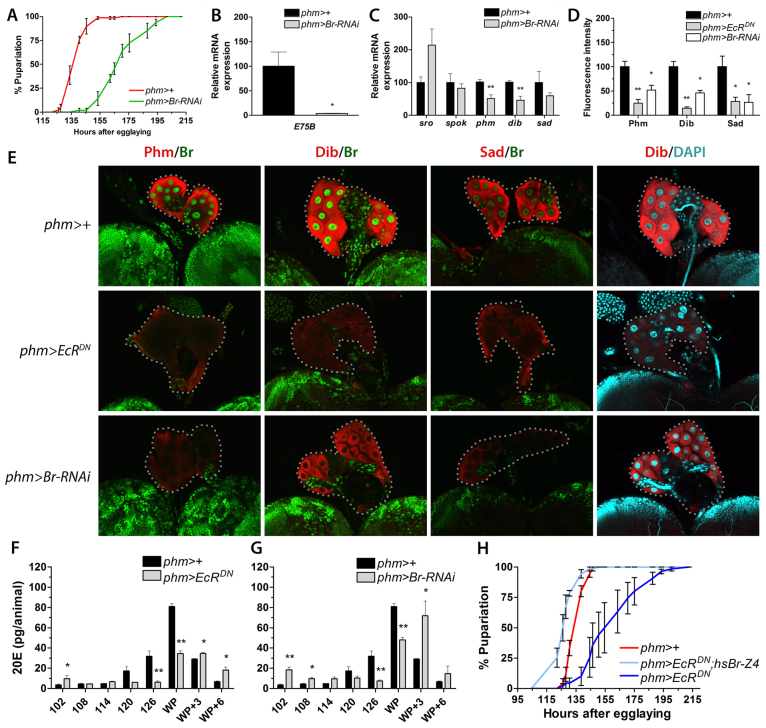
Br is EcR dependent and necessary for the expression of ecdysone biosynthesis genes and the timing of pupariation. (A) Knockdown of br in the PG (phm>Br-RNAi) delays pupariation compared with the control. (B) E75B expression, as a readout for ecdysone signaling, shows that reduction of br expression in the PG (phm>Br-RNAi) results in reduced ecdysone levels in larvae 120 hours AEL (n=4). (C) Transcript levels of genes involved in ecdysone biosynthesis in phm>Br-RNAi larvae 120 hours AEL compared with the control (n=4). (D) Quantification of fluorescence intensity in PG cells (using ImageJ) shows reduced immunostaining of ecdysone biosynthetic enzymes (Phm, Dib and Sad) in the PG of phm>EcRDN and phm>Br-RNAi larvae compared with the control 120 hours AEL. (E) Immunostaining of Phm, Dib or Sad (red) and Br (green) in larval PG cells (120 hours AEL) of the indicated genotypes. DAPI (cyan) in the PG stained for Dib shows that nuclei and cells are intact. Br staining could not be detected in PG cells when EcR activity was reduced (phm>EcRDN) or when br was knocked down in these cells (phm>Br-RNAi). Dotted lines encircle PG cells. All samples were processed and examined using the same microscope settings. Representative images from the PG for each experimental group (n≥5) are shown. (F,G) Ecdysteroid levels in the indicated genotypes during the third instar and prepupal stage. (H) Resupplying Br-Z4 at 112 hours AEL (prior to the high-level ecdysone pulse) by heat-shock treatment of phm>EcRDN; hs-Br-Z4 larvae is sufficient to rescue the developmental delay resulting from PG inactivation of EcR activity. Error bars indicate s.e.m. *P<0.05, **P<0.01, versus the phm>+ control. WP, white puparium.