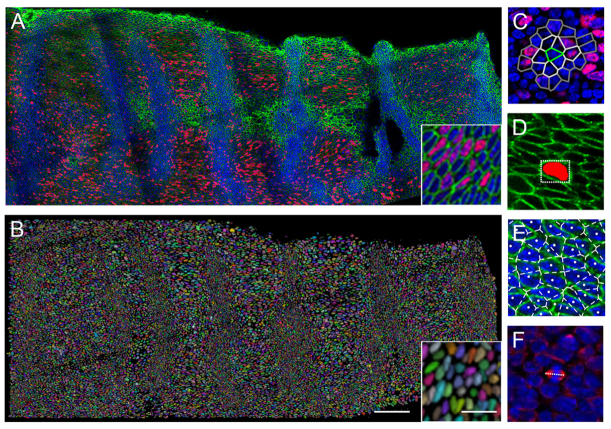
Fig. 2.
Imaging and quantifying cellular parameters. (A) Single z slice from stitched confocal stacks of E14.5 palatal shelf (anterior to the right, medial at the top) using DAPI to label nuclei (blue), and immunofluorescence for BrdU (red) and beta-catenin (green) with detail in inset. (B) 3D rendering of segmented cell volumes in random pseudocolour of the specimen shown in A (inset showing detail). Scale bar: 100 μm; inset, 20 μm. (C) Local BrdU index for a cell (green) includes successive sets of neighbours (shades of grey), shown as Voronoi polygons overlaid on a confocal slice stained for nuclei (blue) and BrdU (red). (D) Cell shape measured from segmented volume (red) by a bounding rectangle at the level of the centroid (white box), shown on a beta-catenin stained (green) confocal slice. (E) Cell territory measured by Voronoi polygons of tessellation of cell centroids (white), shown overlaid on a confocal slice stained for nuclei (blue) and beta-catenin (green). (F) Orientation of mitotic spindle (white line) following immunofluorescence for alpha tubulin marking spindles (red), with DAPI-labelled nuclei (blue).