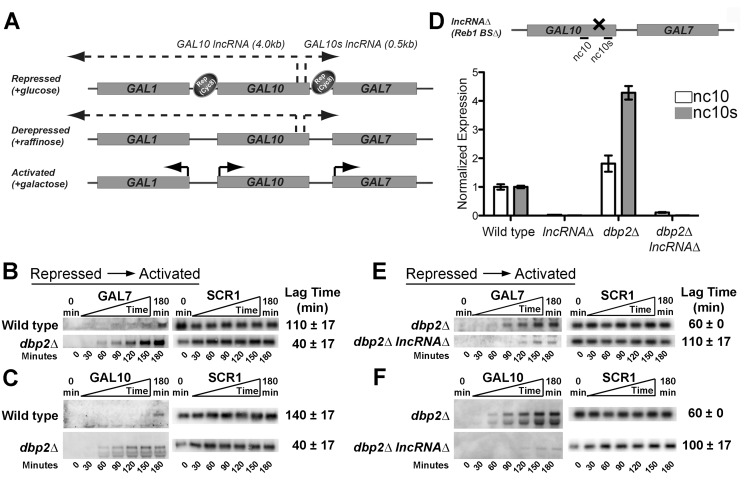
Figure 1. Loss of DBP2 results in rapid, lncRNA-dependent induction of GAL10 and GAL7 from repressed conditions.
(A) Simplified model for carbon-source-dependent regulation of GAL1, GAL7, and GAL10 genes within the GAL cluster. Glucose-dependent repression is mediated by transcription factors (not shown), which then recruit other proteins such as the Tup1–Cyc8 co-repressor complex to promote repression [28], [32], [40], [46], [47], [51]. Derepression occurs under nonrepressing, noninducing conditions when the repressors are no longer present and the GAL genes are not transcriptionally active [29]. Activation only occurs in the presence of galactose [1], [29]. Synthesis of the GAL10 lncRNA, and likely the GAL10s lncRNA, is mutually exclusive with activated expression of the GAL genes [24], [25]. (B–C) GAL7 (B) and GAL10 (C) genes are rapidly induced in dbp2Δ cells following a switch from repressed to activated conditions. Transcriptional induction of wild-type (BY4741) and dbp2Δ strains was conducted by isolating RNA from cells at 30 min intervals prior to and immediately following a nutritional shift from repressive (+glucose) to activated (+galactose) conditions. Transcripts were detected by northern blotting using 32P-labeled, double-stranded (ds)DNA probes corresponding to GAL7, GAL10, or SCR1 RNA as indicated. Each time course was conducted in triplicate. Average lag times to induction are shown with the standard deviation (s.d.) for three, independent biological replicates and correspond to the first time point in a series of time points with increasing GAL transcript levels after normalization to SCR1. An s.d. of zero indicates no variation between biological samples with 30 min time points, whereas an s.d. of 17 indicates a variance of 30 min between replicates. (D, Top) Schematic diagram of the lncRNAΔ strain with GAL10 and GAL10s lncRNAs and primer sets for RT-qPCR. The four previously identified binding sites for the Reb1 transcription factor are present within the 3′ end of the GAL10 coding region [24]. The lncRNAΔ harbors silent mutations that disrupt all binding sites for the Reb1 transcription factor [24]. (D, Bottom) The lncRNAΔ mutation abolishes expression of both the GAL10 and GAL10s lncRNA in wild-type and dbp2Δ cells. GAL10 and GAL10s lncRNAs were detected in the indicated strains following growth in glucose using RT-qPCR as previously described with primers nc10 and nc10s [37]. Transcript levels were normalized to ACT1, which does not vary between these strains, and is the average of three biological replicates with respect to wild type and standard error from the mean (SEM). (E–F) Loss of GAL10 and GAL10s lncRNAs restores repression at GAL7 (E) and GAL10 (F) loci in DBP2-deficient cells. Transcriptional induction assays from repressive conditions were conducted with isogenic dbp2Δ and dbp2Δ lncRNAΔ strains as in Figure 1B–C.