Figure 4.
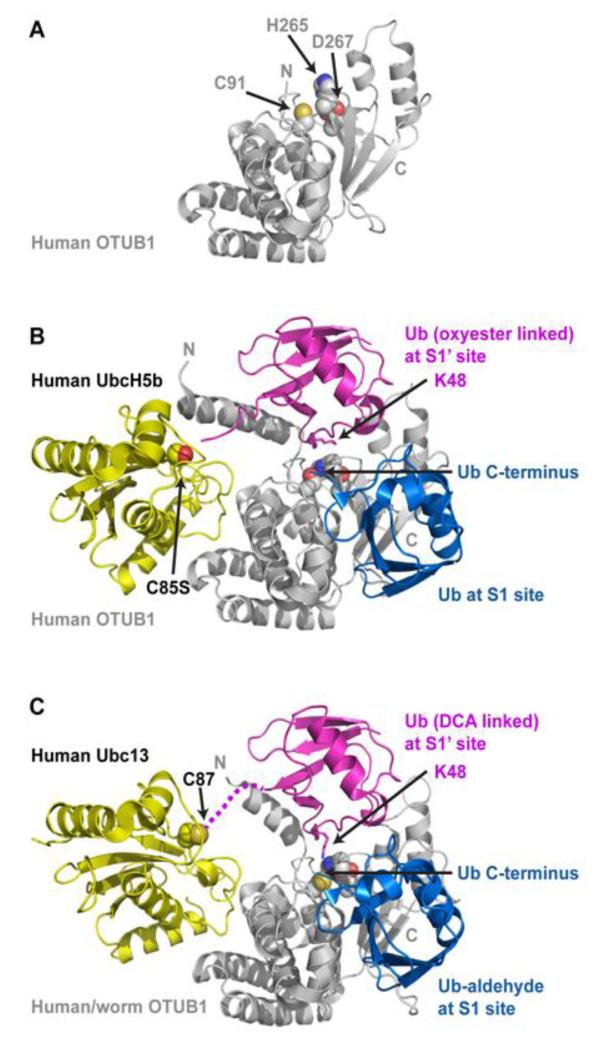
The OTUB1 deubiquitinase binds charged E2s and inhibits their transfer of ubiquitin to E3. (A) The human OTUB1 structure (PDB 2ZFY) with side chains of the catalytic triad (C91, H265, and D267) depicted as spheres. The structure was solved using an OTUB1 deletion lacking the N-terminal 39 residues, which form the N-terminal helix and the S1‘ site. (B) The structure of human OTUB1 bound to UbcH5bC85S-Ub (covalently linked to UbcH5b S85 by an oxyester bond) and free Ub (PDB 4DDI). The E2 UbcH5b and its linked Ub form contacts with the N-terminal helix, and this E2-conjugated Ub binds the S1’ site with K48 positioned towards the catalytic triad and the C-terminus of ubiquitin at the S1 site. (C) The structure of human/worm OTUB1 bound to Ubc13-UbG75C (covalently linked to Ubc13 C87 with dichloroacetone) and Ub-aldehyde (PDB 4DHJ). The hybrid OTUB1 contains the first 45 residues from human OTUB1 followed by the OTU domain from worm OTUB1. Ub aldehyde is covalently bound at the S1 site. OTUB1 binds both Ub-charged E2s in a similar fashion, with K48 of the E2-linked Ub positioned towards the active site and the C-terminus of Ub bound at the S1 site. The binding and arrangement of two ubiquitin molecules at the S1 and S1’ sites in these two structures mimics OTUB1’s natural substrate K48-linked poly-Ub.