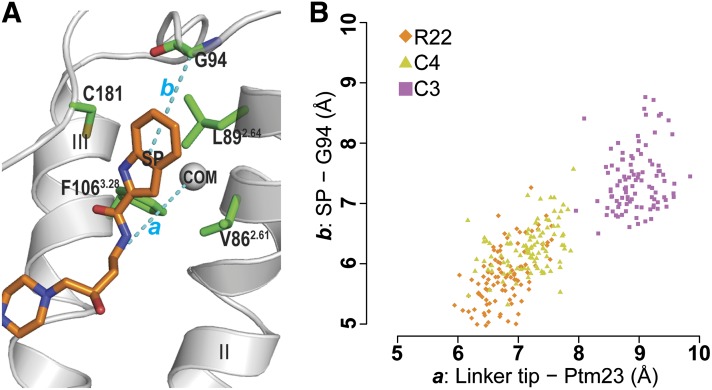
Fig. 3.
Receptor-ligand distances for D3R/R-22, D3R/C4, and D3R/C3. (A) To compare the orientations of the bound ligands in Ptm23 in the MD simulations, distances (cyan dotted lines) were measured between the last carbon atom of the ligand linker region and the center of mass of the Cα atoms of three Ptm23 residues Val2.61, Leu2.64, and Phe3.28 (the gray sphere labeled with “COM”) (a), and between the center of mass of the SP indole ring moiety and the Cα atom of Gly94 (b). The receptor backbone is in gray ribbons, side chains of the Ptm23 residues are in green sticks, and R-22 is in orange sticks. (B) A scatterplot of the receptor-ligand distances, a and b, from the last 2.4 nanoseconds of representative MD trajectories of D3R/R-22 (orange diamonds), D3R/C4 (yellow triangles), and D3R/C3 (magenta squares) shows that the SP moiety of the C3 analog is positioned farther away from the Ptm23 subpocket than those of R-22 and the C4 analog.