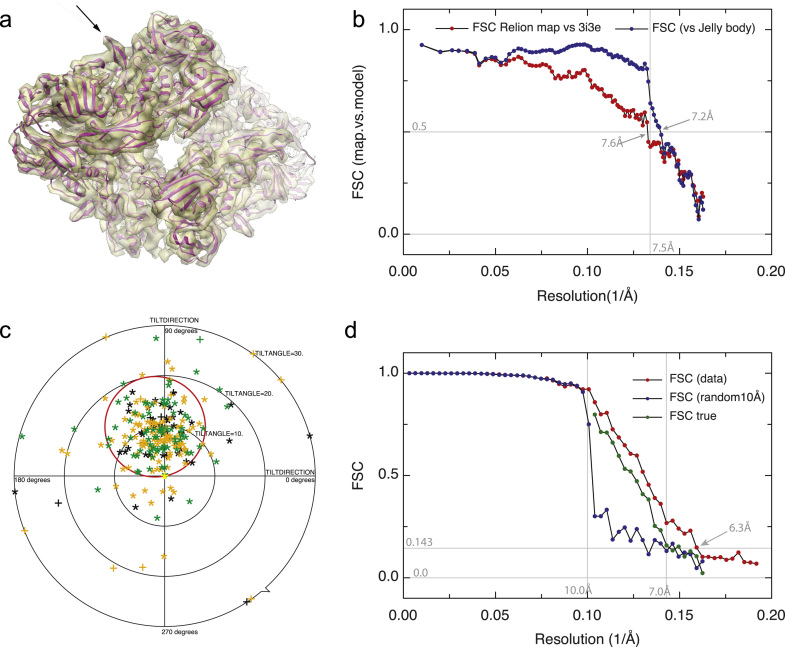
Fig. 5.
Summary of four recommended validation tools. Each panel is from a map chosen to show the value of each tool most clearly. (a) and (b) show comparisons of experimentally determined maps of beta-galactosidase with an atomic model, whereas (c) and (d) show tools that can be used with maps where atomic coordinates are not available. If atomic coordinates are available, then the FSC between map and model provides validation of all steps in the process, although great care is still needed with any flexible fitting procedure. In many cases, however, such as for novel structures at low resolution, atomic coordinates are not available. (a) 3D map of beta-galactosidase from Fig.3(b) obtained using Relion [10] with rigid-body-fitted atomic model superimposed. The arrow shows a beta hairpin. (b) FSC between the Relion map and atomic model. A resolution of 7.6 Å is estimated at 0.5 FSC for a rigid body fit, and 7.2 Å using a jelly body flexible fit to data truncated at 7.5 Å. The value of FSC 0.5 is used in this comparison because the map is calculated from all the images rather only half and the atomic model is assumed to be perfect. (c) Tilt pair parameter plot, which is important for validation at lower (below 1/15 Å−1) resolutions [27]. (d) A typical comparison of the FSC curve (from Fig.3(c)) for a structure with that obtained after HR-noise substitution. In this case, the correlation between the two half data sets shows that about one third of it consists of overfitted noise at 9 Å and half at 7 Å. A genuine resolution of 7.0 Å is estimated from FSCtrue (green symbols), rather than the 6.3 Å value that would be falsely suggested by the overfitted noise (red symbols). The value of FSC 0.143 is used in this comparison because both maps are calculated from only half the images [27] so both contain more noise than the map used for (b). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)