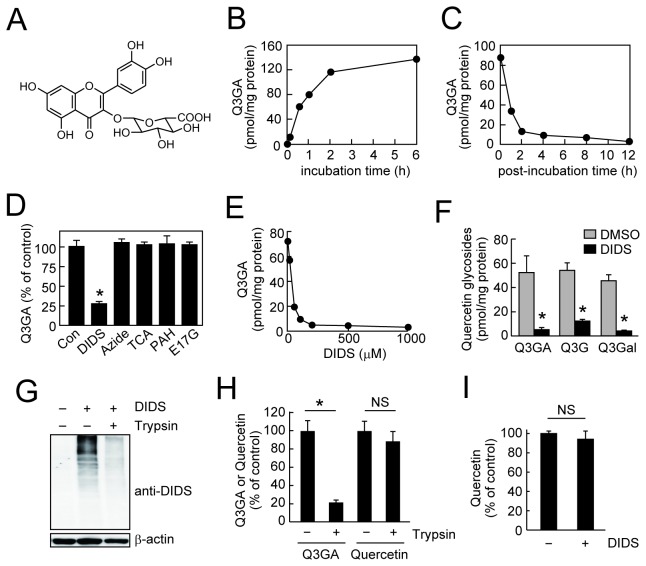
Figure 1. Cell-surface binding of Q3GA.
(A) Chemical structure of quercetin-3-O-glucuronide (Q3GA). (B) Time-dependent accumulation of Q3GA in the RAW264 cells. Cells were treated with Q3GA (50 μM) for the indicated time periods. (C) time-dependent dissociation of Q3GA bound to RAW264 cells. Cells were treated with Q3GA (50 μM) for 1 h, washed, and then incubated in the fresh medium for indicated time periods. Data points represent duplicate determinations. (D) Inhibition of Q3GA accumulation in RAW264 cells by various transporter inhibitors. Cells were pretreated with inhibitors for 15 min, followed by Q3GA (50 μM) treatment for 15 min. The concentrations of inhibitors were as follows: DIDS, taurocholic acid (TCA), p-aminohippuric acid (PAH), and estradiol-17-β-glucuronide (E17G), 500 μM; sodium azide, 5 mM. (E) Dose-dependent inhibition of Q3GA accumulation in RAW264 cells by DIDS. Data points represent duplicate determinations. (F) Inhibition of the accumulation of quercetin-3-O-glycosides in RAW264 cells by DIDS. The DIDS-treated cells were incubated with each quercetin-3-O-glycoside (20 μM) for 15 min. Q3G, quercetin-3-O-glucoside; Q3Gal, quercetin-3-O-galactoside. (G) Anti-DIDS immunoreactivity of the lysates of the DIDS-treated RAW264 cells. Cells were treated with DIDS followed by a quick trypsinization for 1 min. β-Actin was detected as a positive control for intracellular proteins. (H) Effects of quick trypsinization of RAW264 cells on the accumulation of Q3GA and quercetin. Q3GA- or quercetin-treated cells were incubated with trypsin for 1 min. (I) Effect of DIDS treatment on the accumulation of quercetin. Data in all bar graphs are presented as the average ± S.D. (n=3). Asterisks indicate a significant difference (p < 0.05). NS, not statistically significant.