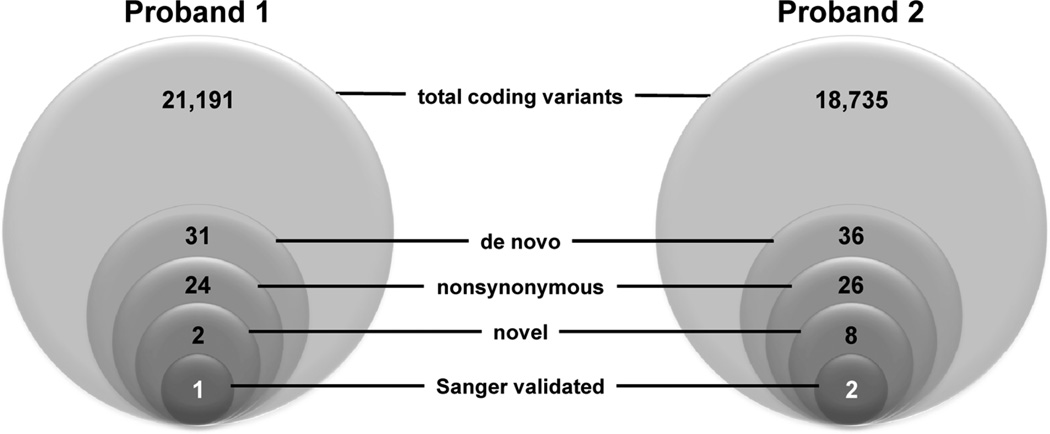
Figure 2.
Exome variant filtering strategy. Sequential filters were applied to the pool of variants discovered by exome sequencing in the two probands. Total coding variants include all synonymous, nonsynonymous, nonsense, frameshift inducing insertions or deletions, and canonical splice site variants within captured exons, but exclude variants in 5’ and 3’ untranslated regions and introns. De novo variants are those found in proband but not in either parent. Novel variants were defined as those absent in public databases (dbSNP, 1000genomes). Novel variants in Proband 1 were also absent in 1202 exomes generated in the Institute of Human Genetics Helmholtz Zentrum München).