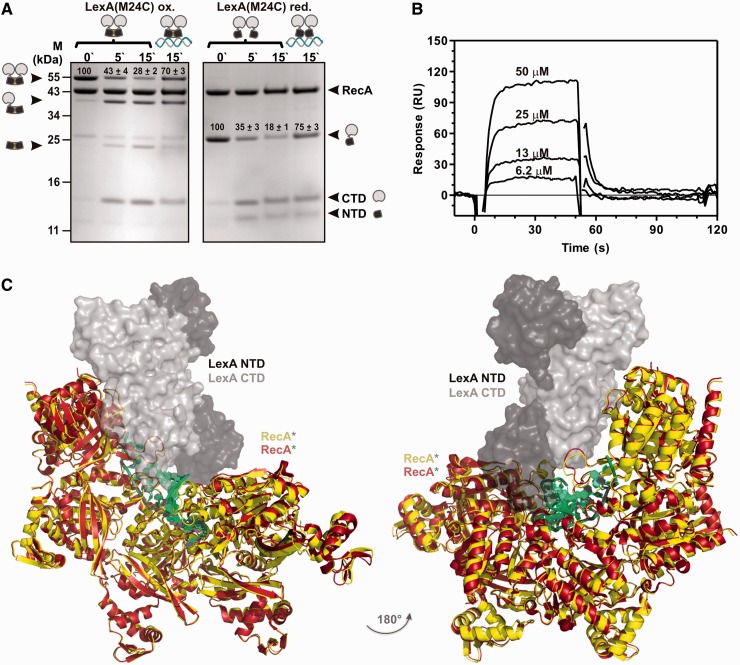
Figure 5.
LexA DNA binding domain interacts with RecA*. (A) LexA in a DNA-binding conformation is inactivated on interaction with RecA*. Time course (min) of RecA*-induced proteolysis of LexA(M24C) in the oxidized, DNA-trapped state, or in the reduced, flexible state. LexA dimer, LexA monomer cross-linked to the N-terminal fragment and cross-linked N-terminal fragments are presented next to the molecular mass marker lane (M). States of the LexA(M24C), the oxidized repressor variant trapped in the DNA-binding conformation (linked NTDs) or the reduced mutant repressor variant interacting with non-specific (marked as LexA without DNA) or the specific DNA (LexA bound to DNA) are presented above the gel. The full length RecA and LexA(M24C) protomers and the cleavage forms of LexA(M24C) (CTD, NTD) are also marked. Quantification of oxidized LexA(M24C) dimers or of reduced LexA(M24C) monomers is presented on the gel above the respective band as the ratio (%) of the protein density value of the initial sample (0 min) relative to the density value obtained from the proteins after 5 or 15 min after addition of RecA*, shown with standard deviation. (B) SPR sensorgram of the binding of the LexA(1–69) NTD with immobilized RecA*. LexA(1–69) in concentration range from 6.2 to 60 µM was injected across the chip-immobilized ∼1000 RU of RecA* for 60 s at 10 µl/min. (C) Secondary DNA of the postsynaptic nucleoprotein filament, enabling exchange of strands between two homologous DNA molecules in recombination, sterically precludes interaction of RecA* with LexA. Postsynaptic RecA* (PDB ID: 3CMX) shown as RecA* in red and dsDNA in green was superimposed by RecA to the LexA–RecA* structure. The RecA* is presented in yellow and LexA in grey (CTDs) and black (NTDs).