Abstract
Translin is a highly conserved RNA- and DNA-binding protein that plays essential roles in eukaryotic cells. Human translin functions as an octamer, but in the octameric crystallographic structure, the residues responsible for nucleic acid binding are not accessible. Moreover, electron microscopy data reveal very different octameric configurations. Consequently, the functional assembly and the mechanism of nucleic acid binding by the protein remain unclear. Here, we present an integrative study combining small-angle X-ray scattering (SAXS), site-directed mutagenesis, biochemical analysis and computational techniques to address these questions. Our data indicate a significant conformational heterogeneity for translin in solution, formed by a lesser-populated compact octameric state resembling the previously solved X-ray structure, and a highly populated open octameric state that had not been previously identified. On the other hand, our SAXS data and computational analyses of translin in complex with the RNA oligonucleotide (GU)12 show that the internal cavity found in the octameric assemblies can accommodate different nucleic acid conformations. According to this model, the nucleic acid binding residues become accessible for binding, which facilitates the entrance of the nucleic acids into the cavity. Our data thus provide a structural basis for the functions that translin performs in RNA metabolism and transport.
INTRODUCTION
Translin, an RNA- and DNA-binding protein, was discovered in extracts of human and mouse cells and shown to assemble into a multimeric complex (1–3). The protein TRAX is a translin paralog found to be specifically associated with translin (4), and also binds single-stranded oligonucleotides when in complex with translin (5). Both translin and TRAX are highly conserved throughout eukaryotic evolution. Thus, homologs of both proteins are found in many eukaryotes, from Homo sapiens to Drosophila to the fission yeast Schizosaccharomyces pombe (6).
Translin and the related protein TRAX appear to be multifunctional. Based on its preferential affinity for specific sequences of single-stranded DNA flanking chromosome translocations as well as single-stranded microsatellite repeats and G-strand telomeric repeats, translin has been proposed to be involved in different functions of DNA metabolism (7–9). More importantly, the interaction of translin and translin–TRAX complexes with RNA might be involved in the control of mRNA translation and transport (2,10–14). More recent studies provide evidence for the involvement of translin–TRAX complexes in RNA interference (15–18) and tRNA processing (19). In summary, translin and translin–TRAX complexes play an important role in eukaryotic cell metabolism involving the processing of single-stranded nucleic acids.
Although dimers of translin were proposed to be the minimal unit that can bind single-stranded RNA and DNA in vitro (20), biochemical studies have shown that the major binding configuration of the protein is a homo-octamer (7). The first detailed electron microscopic (EM) analyses of recombinant translin suggested that it formed octameric ring structures with eight-fold (C8) symmetry axis and a central open channel that was suggested to accommodate single-stranded oligonucleotides (21,22). In their quaternary organization and subunit dimerization, these proposed translin rings appeared to be similar to hexameric ring helicases (23,24). However, x-ray crystallography analyses of the virtually identical mouse and human translin (99% sequence identity) revealed a different octameric assembly, consisting of a dimer of tetramers with two symmetry axes: one two-fold (C2) axis relating the two tetramers, and one four-fold (C4) axis relating the four dimers (25,26). The human translin–TRAX complex also showed a similar X-ray structure, with the same dimer of tetramers architecture in which two protomers of the octameric translin had been exchanged by TRAX (17). Interestingly, the recently published crystallographic structure of Drosophila translin showed the same asymmetric assembly observed in human translin–TRAX (27). However, in a different study, EM reconstructions of Drosophila translin–TRAX, while keeping the overall dimer of tetramers organization, revealed significant conformational rearrangement with a large expansion from the X-ray structure (18).
Our previous work identified and mapped oligonucleotide-binding interfaces on individual subunits of pure translin octamers (28). These interfaces were located within a large internal cavity in the crystallographic structure of translin, which in principle could accommodate nucleic acids. However, such cavity is not accessible for nucleic acid binding in the crystallographic structure, while it would be accessible in other octameric configurations such as the ones suggested by the translin and translin–TRAX EM data (18,22). Thus, several key questions regarding the structural basis of translin function remain open. Which of the above described translin octameric structures correspond to the stable translin configuration in solution? Are any of these structures able to bind nucleic acids? Are there other conformational rearrangements of translin which could allow the functional binding of nucleic acids?
In an attempt to furnish answers to these questions, we have performed an integrative study that provides new functional structures for translin and translin–RNA complexes in solution. In this work, we combined small-angle X-ray scattering (SAXS) measurements and site-directed mutagenesis experiments with computational approaches. Our data demonstrate that the configuration of translin and translin–RNA complexes in solution is a dimer of tetramers, which undergoes conformational transitions that had not been previously identified. This conformational heterogeneity clarifies the mechanism for nucleic acid binding by translin and provides a structural and dynamic picture that explains its biological functions.
MATERIALS AND METHODS
Preparation of recombinant human translin
A translin-encoding plasmid was transfected into the Escherichia coli XL1 bacteria. Translin was purified from the transformed bacteria as described (8) with some modifications. Briefly, a bacterial culture of 200 ml was grown to late log phase in LB medium containing 0.05 mg/ml of ampicillin. At this stage, isopropyl-β-d-thiogalactopyranoside (IPTG) was added at 2 mM and the culture was further incubated for 3 h. The bacteria were centrifuged, and the bacterial pellet was resuspended in 10 ml of phosphate buffer (3.4 mM NaH2PO4, 46.6 mM Na2HPO4, 300 mM NaCl; pH 8.0) containing 25 mM imidazole and complete EDTA-free mini protease inhibitor cocktail tablets (Roche), and the bacteria were lysed in a French pressure cell press. The lysate was centrifuged and the supernatant was purified in a Ni-agarose column, as described (8) with the following modifications: after loading of the lysate, the column was washed successively with 30 ml of phosphate buffer containing 25 mM imidazole and with 12 ml of phosphate buffer containing 45 mM imidazole. Then, translin was eluted in 5 ml of phosphate buffer containing 250 mM imidazole. The purified translin was dialyzed against 1000 ml of 100 mM NaCl, 10 mM Tris-Cl pH 8.0, 0.1 mM EDTA, 2 mM DTT and 5% glycerol. The molecular weight of the purified translin protomer is 26.8 KDa, corresponding to 240 aminoacids including 12 from the His-tag. The concentration of translin was determined by Coomassie staining using Coomassie Protein Assay Reagent (Bio-Rad) against BSA standard. Alternatively, the concentration of the protein was determined by measurements of absorption of UV light at a wavelength of 280 nm using Nanodrop. The protein concentrations were calculated from the absorption values using the corresponding extinction coefficient of 16 750 M−1cm−1 at 280 nm (8). The concentrations of translin determined by the staining procedure and by UV absorption measurements were found to be equivalent (±10%).
Site-directed mutagenesis
Mutations in plasmids encoding translin were generated by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The sequence of the whole translin gene was determined in all mutants to ensure that mutations only occurred in the desired locations. Oligonucleotides were purchased from Sigma and from MWG-Biotech AG. The plasmid expressing translin was kindly sent to us by Dr M. Kasai from the Department of Immunology, National Institutes of Health, Tokyo, Japan. This construct was made by cloning the translin cDNA into the plasmid pQE-9 (Qiagen), such that the recombinant protein expressed from this plasmid in E.coli bacteria includes a His6 tag at the N terminus (8).
Glycerol gradient centrifugation
Recombinant translin was loaded on a 10 ml linear 20–40% (v/v) glycerol gradient containing 10 mM Tris–HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA and 2 mM DTT. The tubes were centrifuged for 24 h in a Beckman ultracentrifuge in the SW-41 rotor at 38 000 rpm at 8°C. Fractions of 0.22 ml were collected from the bottom of the gradient. The position attained by the protein was determined by SDS–PAGE of aliquots withdrawn from each fraction, followed by Coomassie blue staining of the gel. Estimations of the sedimentation constant and the molecular weight of translin were obtained by comparison with sedimentation constants of known protein markers, as previously described (29,30). The glycerol gradient centrifugation experiments performed on translin wild-type confirm the monodispersity of the preparation corresponding to an octameric arrangement.
Small-angle X-ray scattering experiments
The SAXS measurements for translin and translin–RNA complex were carried out at the EMBL BioSAXS beamline X33 at the DORIS Storage Ring, DESY (Hamburg, Germany) using an X-ray wavelength of 1.54 Å and a sample-to-detector distance of 2.7 m. Translin samples were measured at 10°C at concentrations of 5.9, 4.1, 3.0, 1.5 and 0.8 mg/ml in a buffer containing 10 mM Tris–HCl, pH 8.0, 0.1 mM EDTA, 1 mM DTT, 5% glycerol and 100 mM NaCl. Translin–RNA complexes were prepared by mixing in the same buffer translin octamers with the RNA oligonucleotides at a stoichiometric ratio of 1:1.5, followed by incubation at 25°C for 20 min. SAXS measurements of these complexes at protein concentrations of 5.9, 3.0 and 1.5 mg/ml were also carried out at 10°C. We have previously found that by following the above mentioned incubation at 25°C, cooling of translin–nucleic acid complexes to 10°C did not affect the stability of the complexes (data not shown). The scattering patterns of the corresponding buffer solutions were recorded before and after the measurements of the protein sample. The scattering profiles measured covered a momentum transfer range of 0.0087 < s < 0.60 Å−1. Inspection of the consecutive 30-s X-ray exposures discarded the presence of radiation damage. No signature of aggregation was observed in any of the curves. In addition, no noticeable concentration effects were observed in the datasets indicating the absence of oligomeric equilibrium (Supplementary Figure S1). Inspection of the low-angle region of the curves indicates the presence of attractive interparticle interactions (Supplementary Figure S1) that were minimized when merging the curves (see below). Final curves at each concentration were derived after the averaged buffer patterns were subtracted from the protein ones using standard protocols with PRIMUS (31). Analyzed curves for translin and translin–RNA samples were obtained by merging the curves at different concentration using standard protocols to keep the maximum sensitivity while avoiding inter-particle interactions. Molecular weight estimations for translin and translin–ssRNA from SAXS data were performed using Porod’s volume approach and the program SAXS-MoW (32). From Porod’s volume, molecular weight estimates for translin and translin–ssRNA were 253 and 270 KDa, respectively, using a value of 1.6 for transformation. When using SAXS-MoW up to 0.3 Å−1 MW estimates of 261 and 273 KDa were obtained for translin and translin–ssRNA, respectively. In all these cases the MW is overestimated with respect to the theoretical one for a translin octamer (220 KDa). This disagreement can be explained by the fact that translin is not a globular protein but a hollow sphere. The forward scattering, I(0), and the radius of gyration, Rg, were evaluated using the Guinier's approximation (33), assuming that at very small angles (s < 1.3/Rg), the intensity can be well represented as I(s)=I(0) exp(−(sRg)2/3). The maximum particle dimensions, Dmax, and the pair-wise distance distribution function, p(r), were computed with the program GNOM (34) using a momentum transfer range of 0.020 < s < 0.30 Å−1. The agreement between the SAXS curve and the crystallographic structure was evaluated with CRYSOL (35) using a momentum transfer range of 0.020 < s < 0.40 Å−1.
Modeling of full-length translin
The N-terminal histidine-tag and the C-terminal tail were absent from the human translin crystal structure; hence, they were assumed to be disordered in solution. We used the program RanCh (36) to calculate 10 random conformations for the N-terminal histidine-tag and C-terminal tail of 12 and 11 residues, respectively, per each of the eight subunits of the crystallographic structure (PDB 1J1J). We used these sets of conformations to reconstruct these terminal tails for all assemblies calculated along the present study by superimposing the Cα atoms of the individual tails onto the equivalent ones in each corresponding translin subunit.
Translin conformational sampling by normal mode analysis
We have explored the conformational space of human translin (PDB code 1J1J) based on normal mode analysis. Normal Mode Analysis (NMA) assumes that the protein structure is a system of coupled harmonic oscillators connecting the atoms (to simplify calculations, only Cα atoms are often considered). The dynamics of such a system is constituted by the normal modes, each one involving all the particles of the system. Based on the normal modes that produce the most relevant conformational movements (usually the lowest frequency ones), taken either as single modes or in combination, one can reproduce (at least partially) the conformational heterogeneity in a protein. For this, we built translin models by combining three different NMA-based approaches, to explore more extensively the conformational space of translin. All NMA calculations started from the Cα coordinates of translin, excluding the unstructured N-terminal histidine-tag and C-terminal tail. First, we used elNémo server (http://www.igs.cnrs-mrs.fr/elnemo/) (37) to exhaustively obtain displacements using the five lowest-frequency modes, either as individual or by combining them in pairs, with amplitude perturbation between 2000 and −2000 and step size of 100 (arbitrary units). This generated 1425 models. After removing highly distorted models, i.e. with more than 200 pairs of consecutive Cα atoms with distance artificially larger than 5 Å, we obtained a final number of 1415 models. Secondly, we used a Monte-Carlo protocol (38,39) to sample conformations based on combinations of the five lowest-frequency modes with random amplitudes. We adjusted the internal parameters to produce models with similar amplitude perturbations to those obtained by elNémo. To generate the same number of conformations, we initially built 3000 models, removed highly distorted models, and randomly selected 1415 models. Finally, we used iMC module from iMOD (40) to sample conformations by randomly selecting one of the five lowest-frequency modes in internal coordinates and its amplitude, in an iterative process. We applied the default parameters and produced the same number of models as the other two methods (1415) for the sake of consistency. The results from the three normal mode sampling methods produced a final set of 4245 Cα models, which were rebuilt by adding missing atoms and side chains and atomically refined with Modeller 8v1 (using the original Cα model as template) to fix incorrect bond distances (41,42).
Structural modeling of translin–RNA complexes
We have devised here a new computational method for flexible docking of the single-stranded RNA to translin, based on the identification of possible dinucleotide binding sites on the translin surface to build different conformations of RNA bound to translin (Supplementary Figure S6). More specifically, we used FTDock v2.0 (43) to dock a rigid GU dinucleotide [modeled by MMB v2.3 (44)] to all the conformations found in the SAXS analysis of the free translin, as described in next sub-section and explained with more details in the Results section (as well as to the crystallographic structure, PDB code 1J1J). For the sake of efficiency, we selected only dinucleotide poses that were within 5 Å inter-atomic distance from any of the residues in the known nucleic acid binding equatorial region (R21, R36, R86, R92, Q96 and R192), as previously defined (28). The resulting GU docking poses were then clustered using a cutoff of 10 Å inter-atomic distance, and for each cluster, the GU docking pose with the best FTDock SCscore was selected as representative. All possible combinations of up to 12 consecutive dinucleotide docking poses were calculated using a graph-like algorithm, allowing “gaps” that would represent possible regions of the RNA molecule that would not be bound to translin. We clustered those combinations that were >70% identical (i.e. at least 9 out of the 12 dinucleotide docking poses were the same). Finally, for each representative combination, a helicoidal 5′-GUGUGUGUGUGUGUGUGUGUGUGU-3′ ssRNA molecule was modeled by MMB v2.3 (44) and refined by a quasi-Newton optimization protocol restrained to the C4' atom positions of the dinucleotide docked poses. This procedure resulted in a large initial list of 65 982 translin–RNA complex structures that included many impossible models with excessive number of atomic clashes with translin, as no specific conformation was imposed to the ssRNA when it was threaded to the docked dinucleotides. Therefore, we removed models with >150 atomic clashes and obtained a final list of 2865 translin–ssRNA conformations.
Fitting structural models to SAXS data
The program CRYSOL (35) was used to calculate the theoretical SAXS curves for each of the full-length translin and translin–RNA models. Default parameters for CRYSOL calculations were used to derive theoretical profiles. All theoretical curves were computed with a number of 201 points and a maximum scattering vector of 0.4. For each octameric assembly, 10 different structures were computed by random addition of terminal tails to realistically cover the conformational sampling (see above). The individual theoretical curves were averaged and compared with the appropriate experimental SAXS curve, using χi as a figure of merit in a momentum transfer range of 0.020 < s < 0.40 Å−1.
In the case of free-translin, Minimal Ensemble Search (MES) (45) was used to find a minimum ensemble of conformations from the 4245 models built with NMA (in which terminal tails were modeled as explained above). In the case of translin–RNA complex, we calculated the SAXS profile for all generated translin–RNA models (see above), including the disordered N- and the C-terminal regions of translin, and evaluated which ensembles of conformational states provided the best fitting to the SAXS data, using MES for combinations of up to five conformational states (45). To further study the conformational heterogeneity of ssRNA within translin, we also used the program Ensemble Optimization Method (EOM) (36) for combinations of up to 50 conformational states (five different runs were performed with EOM allowing structure repetition).
RESULTS
Translin is a dimer of tetramers in solution
To clarify the 3D arrangement of translin in solution and the structural basis for its nucleic acids recognition, we have studied the structure of human translin by SAXS, which provides information about the overall size and shape of biomolecules in solution (46–48). Analysis of the low-resolution part of the SAXS curve using Guinier’s approximation (33) indicates that translin is a particle with a radius of gyration (Rg) of 48.4 ± 0.4 Å in solution (Supplementary Figure S1). The pairwise distance distribution function, p(r), as derived from the SAXS curve, presents a prominent peak at ∼58 Å indicating that translin adopts a globular structure in solution (Supplementary Figure S1). In addition, the p(r) function displays a smooth decrease to the maximum intra-particle distance, Dmax, of 160 ± 5 Å. The overall picture derived from the primary analysis of the raw data would favor the C4 octameric model found in the crystallographic analyses of translin and translin–TRAX complexes as well as in the EM study of translin–TRAX (17,18,25,26), but is not compatible with the C8 octameric ring model proposed in an earlier EM study of translin (22). We further confirmed this observation by fitting of the C4 and C8 structural models to the SAXS data. These calculations also indicated that only the C4 octameric translin found by X-ray crystallography (1J1J PDB) displayed a reasonable agreement to the SAXS experimental curve (Supplementary Figure S2 and Supplementary methods).
Translin shows conformation variability
The above-described SAXS data clearly favor the compact translin C4 arrangement derived by X-ray crystallography (25,26). Nevertheless, despite this overall agreement, the fitting was not optimal (χi = 7.19). Two major reasons for this suboptimal fitting can be hypothesized: (i) the highly flexible N-terminal histidine-tag and the C-terminal tail, of 12 and 11 residues, respectively, which are present in the sequence construct used for experiment, but missing in the X-ray structure; and (ii) a putative conformational change that, while keeping the octameric C4 arrangement, could substantially modify the overall shape of the translin complex as suggested by the EM structure of Drosophila translin–TRAX (18).
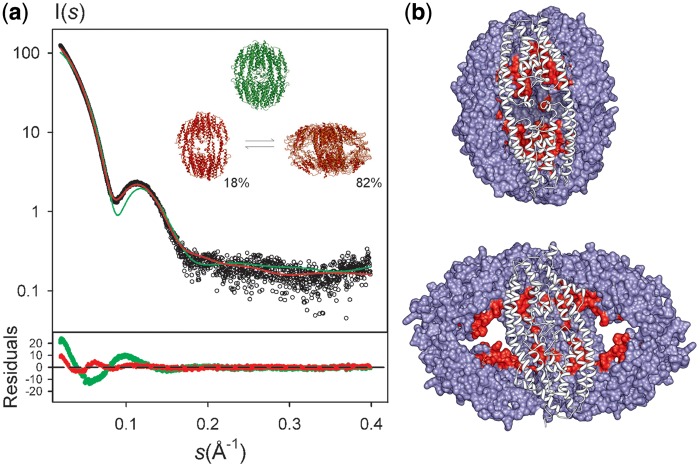
The presence of disordered tails is compatible with the smooth decrease of p(r) towards the Dmax value of 160 ± 5 Å. This hypothesis was tested by building models for full-length translin including the terminal tails (see ‘Materials and Methods’ section and Supplementary Figure S3). The averaged curve from an ensemble of 10 full-length translin conformers having the same global structure, but with terminal tails in random conformations, was compared with the experimental one (Figure 1a). Although the presence of the disordered tails improved the fit to the SAXS data, the goodness of fit value was not yet satisfactory (χi = 5.01), suggesting that translin may adopt a global conformation in solution that differs from that in the crystal.
Figure 1.
Structural models of translin according to SAXS data. (a) Experimental fit of the structural models of full-length translin (green) and best MES dynamic ensemble (red) to the experimental SAXS curve (black circles). The structure of the crystallographic translin yielded a fitting to SAXS data of χi = 5.01. The best ensemble of translin conformers was found to be formed by a compact and an ensemble of three open conformations (with individual relative populations of 45.8, 23.3 and 13.3%), with a fitting to SAXS data of χi = 1.67. Point-by-point deviations for each model are displayed at the bottom of the plot with the same color code. Although the full-length protein has been used in both fittings, flexible N-terminal histidine-tag and C- terminal regions are not shown here for the sake of clarity. (b) The RNA/DNA binding residues according to OPRA (49) prediction (previously validated in S.Pombe (28)) are shown in red for the two major configurations of translin.
The global conformational space of translin was explored by NMA, a powerful technique that allows efficient conformational sampling in cases of large structural arrangements (50). We generated 4245 models of the translin octamer, and for each of them, a theoretical SAXS curve was computed by averaging the data derived from 10 different conformations of the full model including the N-terminal histidine-tag and C-terminal tail (‘Materials and Methods’ section and Supplementary Figure S4). The best fitting obtained by a single model was notably better than that obtained by the crystallographic structure (χi = 2.24), but still far from optimal, suggesting that there could be conformational heterogeneity in solution. Therefore, an optimal combination of a small number of these models/curves that collectively describes the SAXS curve was obtained using the program MES (45) (see Materials and Methods). The conformational equilibrium found for translin notably improved the fitting to the SAXS curve (χi = 1.67, see Figure 1a) by combining one compact conformation (18% population) and an ensemble of three open conformations (82% population). The compact conformation is very similar to the structure obtained by crystallography, but slightly more open in the longitudinal interdimeric region (Supplementary Figure S5). Open conformations present a maximum interparticle distance of 152.0 Å, in excellent agreement with the experimentally determined Dmax, 160 ± 5 Å. Interestingly, the open conformations found in this analysis leave the nucleic acid-binding residues accessible for interaction (Figure 1b).
Site-directed mutagenesis supports the dynamic octameric model of translin
To further examine the proposed oligomerization model for translin, we generated a series of amino acid substitutions in the protein by site-directed mutagenesis, chosen according to the expected protein–protein interfaces in the different octameric models proposed for translin. These mutated translin variants were designated Nt-1 (N-terminal 1), Ct-1 (C-terminal 1), Ct-2 (C-terminal 2), Nt+Ct and Equatorial, named according to the positions of the mutations in the monomer (Table 1; Figure 2). The translin protein variants encoded by these mutants were purified and their major oligomeric states were determined by ultracentrifugation in glycerol gradients (Figure 2a). It can be seen that while the wild-type translin sedimented as octamers, the major form of the protein variant Nt-1 sedimented as tetramers. It is also seen that the variants Ct-1 and Ct-2 sedimented as dimers, while the variant Nt+Ct sedimented as monomers.
Table 1.
Summary of properties of wild-type translin and of translin variants
| Translin | Mutations | S. value | MW (kDa)a | No. of subunitsb |
|---|---|---|---|---|
| Wild type | 9.11 ± 0.41 | 220.02 | 8 (8.2) | |
| Nt-1 | I6A/F7A | 5.23 ± 0.24 | 90.83 | 4 (3.9) |
| Ct-1 | E206A | 3.88 ± 0.24 | 56.43 | 2 (2.1) |
| Ct-2 | R153A | 3.81 ± 0.27 | 55.41 | 2 (2.0) |
| Nt+Ct | I6A/F7A/E206A | 2.77 ± 0.12 | 32.33 | 1 (1.2) |
| Equatorial mutant | Y85A/R129A,/ R182G/L183V | 8.98 ± 0.52 | 215.24 | 8 (8.0) |
aThe molecular weights were calculated from the average S values as described in ‘Materials and Methods’ section.
bEstimates of the number of subunits (in brackets) were obtained by dividing the experimentally determined molecular weights of the complexes by the molecular weight of the human translin subunit (26.8 KDa). The actual number of subunits is also indicated. An octamer of translin has theoretical MW of 215 KDa.
Figure 2.

Glycerol gradient centrifugation of wild-type and mutant translin variants. (a) Sedimentation profiles of recombinant wild-type translin and the mutant translin variants listed in Table 1. Vertical arrows indicate the peaks of the major configuration of each of the translin mutant variants. (b) The two adjacent substitutions I6A and F7A of the mutant Nt-1 break the octamer in tetramers (a tetramer is shown in orange in the compact and open translin models, with the corresponding mutated residues in red CPK). The insert shows a hydrophobic pocket in which I6 and F7 of one monomer (in cyan) interact with the opposing residues of the second monomer (in orange). (c) The E206A substitution of Ct-1 mutant or the R153A substitution of Ct-2 break the octamer into dimers (one dimer is shown in orange in the compact and open translin models, with the corresponding mutated residues E206 in red CPK and R153 in blue CPK). The insert shows an ion bridge (indicated with two straight lines) between E206 in one monomer (in cyan) and R153 of the second monomer (in orange). (d) The Nt+Ct mutations (I6A, F7A and E206A substitutions) break the translin octamer into monomers (a monomer is shown in orange, with the corresponding mutated residues in CPK). (e) The Equatorial mutant (R182G/L183V/R129G/Y85A) has no effect in the octameric structure. Interestingly, according to the crystallographic structure, these mutants in a given monomer (in orange) are interacting with other monomers, while in the open conformation, these mutants are not located at the oligomer interface, which explains the incapability of this mutant to break up the octamer (see Discussion).
Figure 2b–d shows the positions of these mutations at the interfaces of the open and closed octameric structures. Figure 2b shows the two substituted amino acids, I6 and F7 of each subunit in the variant Nt-1. These two hydrophobic amino acids in the α-helix 1 of each subunit interact with a cluster of hydrophobic amino acids in α-helices 1, 2, 3 and 4 of the adjacent subunit (see inset in Figure 2b). Hence, the substitutions of I6 and F7 by alanine were expected to cause disruption of this interaction and the separation of the two tetramers along the longitudinal axis, as observed. A similar representation of the E206A and R153A substitutions in the variants Ct-1 and Ct-2 is illustrated in Figure 2c, depicting a salt bridge formed between the residues E206 and R153 in two adjacent subunits (see inset in Figure 2c). Either of the two substitutions E206A or R153A disrupted this strong interaction, thereby causing the separation of dimers along the transversal axis. As expected, combining Nt-1 and Ct-1 substitutions in the Nt+Ct variant led to disruptions along both axes and produced monomers (Figure 2d).
Finally, the Equatorial four-mutant construct (Y85A/R129A/R182G/L183V) harboring changes along the equatorial axes of the monomer did not disrupt the octameric arrangement (Figure 2a). Interestingly, these substituted amino acids are (albeit weakly) involved in the interactions between the subunits in the X-ray-based octameric structure (Figure 2e), but they are not at all involved in the interactions between subunits in the open conformations. Thus, the observation that these equatorial mutations did not disrupt the octameric arrangement is consistent with the existence of highly populated open conformations in solution.
RNA is bound to the central cavity of the dynamic translin assembly
To identify the structural features of RNA recognition by translin, SAXS data were obtained for a translin–RNA complex formed by the mixing of translin and the oligoribonucleotide (GU)12, which has high affinity for translin (Kd = 2 nM) (8,29). The SAXS curve measured for this complex displayed similar features to that of the free translin, indicating that the overall arrangement is not changed upon ssRNA binding. The nucleoprotein complex presents a Rg value of 47.2 ± 0.3 Å that is slightly smaller than that found for free translin, 48.4 ± 0.4 Å (Supplementary Figure S1). The p(r) function derived from the SAXS curve of the complex presents an equivalent Dmax value to that found for free translin, 160 ± 5 Å, indicating that similarly open conformations are present in the complex. The p(r) function of the complex presents a maximum value at a slightly smaller radius, 56 Å, than that of free translin, 58 Å. This last observation and the smaller Rg value found in the complex can be explained if the ssRNA is placed in the interior of the translin cavity, partially filling the hollow sphere that represents the protein (46).
To have a more detailed picture of the translin–RNA complex, we generated putative structural models by applying an innovative procedure for flexible ssRNA docking. The X-ray structure of translin and the four conformations derived from the SAXS analysis of the free translin were used as receptors to dock rigid GU dinucleotides that were subsequently connected using a graph-like algorithm to create 2865 models of the nucleoprotein complex (see Materials and Methods and Supplementary Figure S6 for details). We tested the capacity of each one of these models to describe the experimental curve. The best fit obtained (χi = 2.44) is displayed in Figure 3 and presents systematic departures from the experimental curve in a broad momentum transfer range. The incapacity to describe the experimental curve with a single conformation prompted us to use ensemble methods. Using MES as optimization tool, several ensembles of translin–RNA complexes were found with a similarly good agreement to the SAXS data (χi = 1.73–1.75) (Supplementary Figure S7). In all these ensembles, the relative population of compact (10–16%) and open (84–90%) translin forms was similar to that in the free translin, consistent with the equivalent Dmax values found in both cases (Dmax, 160 ± 5 Å). These values are also in excellent agreement with the maximum interparticle distance of 152.8 Å calculated for the open translin forms in these complexes. The ssRNA molecules in these complexes adopted many different conformational states (Supplementary Figure S7). However, it is not possible to disregard that such conformational heterogeneity might arise from the low-resolution nature of the SAXS technique, which does not allow distinguishing between some of the different RNA conformers generated during the docking process. To further explore the conformational heterogeneity of bound RNA, we evaluated the fitting to the SAXS curve by using the EOM, which allows the consideration of larger ensembles of coexisting structures (up to 50) (36). An excellent agreement to the experimental curve was obtained (χi = 1.62), and the relative population of open (22%) and compact (78%) translin forms was similar to that previously found with MES for the complex and the free states (Figure 3a). The ssRNA molecules found in the optimized ensemble showed a large conformational heterogeneity, which is compatible with a model in which the RNA molecules do not adopt a fix configuration when placed within the translin cavity (Figure 3b). In a scenario where translin displays a high level of plasticity, an intrinsically flexible molecule such as ssRNA, which is composed of a repetitive sequence and lacks a localized recognition region, most probably may adopt multiple configurations within a symmetric cavity hosting multiple binding sites.
Figure 3.
Structural and dynamic models of translin–ssRNA complexes according to SAXS data. (a) Experimental fit of the best single-conformation of the complex (in green) and the dynamic structural model of translin–RNA interaction (in red) to the experimental SAXS curve (black circles). Point-by-point deviations for each model are displayed at the bottom with the same color code. (b) The best equilibrium of conformers was found to be formed by an ensemble of compact and open translin forms bound to structurally different RNA molecules, with a fitting to SAXS data of χi = 1.62, notably better than the one found for the best single conformation (χi = 2.44). Translin is shown in gray ribbon, while the docked RNA conformations are shown in ribbon and surface, colored according to their population within the global ensemble (red is higher population; blue is lower). Two different views are displayed.
DISCUSSION
Translin is a highly flexible assembly in solution
Our results clearly demonstrate that translin forms a highly plastic octameric assembly in solution. This flexibility was already suggested by the absence of conserved hydrogen bonds between the two tetramers in the crystallographic structure of human translin (26). Here, we present SAXS data that actually indicate the coexistence of two major arrangements of translin octamers in solution: open and compact. The open state constitutes ∼82% of the octamers population, whereas the compact state only represents ∼18% of the octamers. We should note here that the presence of residual interparticle interactions not completely suppressed by the data processing (see Methods) could entangle the exact determination of the relative population of both families of conformations, causing a slight overestimation of the fraction of molecules in the open conformation. The compact form is structurally similar to the crystallographic structure, indicating that during the crystallization process the more symmetrical and compact form is selected despite not being the major species in solution.
The conformational changes of translin involve the loss or gain of most of the dimer–dimer interactions, which suggests that these are formed by weak intermolecular contacts. This model explains why our Equatorial variant remains octameric, as the mutated residues are not involved in dimer–dimer contacts in the open state (Figure 2e). In contrast, strong interdimeric interactions occur in the N-term and C-term regions of translin in both the open and compact models. These contacts maintain the structure of the global assembly while allowing the conformational dynamics of the system. As a consequence, mutations in these regions cause disassembly of the translin octamer into different oligomeric states (Figure 2b–d).
These observations regarding the inherent plasticity of translin are consistent with the Debye–Waller factors (B-factors) obtained in the crystallographic refinement of translin (PDB 1J1J) (26), which displays an inhomogeneous behavior along the sequence. Interestingly, residues in the equatorial region of the structure, located at the hinge of the dimeric units, present larger B-factors, suggesting an increased degree of mobility in comparison with residues in the polar regions (Supplementary Figure S8). The conformational fluctuations found in the crystallographic environment are compatible with the necessity of flexibility in those regions, which allows the transition between the open and compact translin conformations in solution. It remains to be seen whether structurally homologous systems, like translin–TRAX octameric complex, also have the same alternative conformational states in solution as pure translin octamer. In view of the high homology of translin and TRAX, one would expect similar structural architecture. However, the presence of one or several catalytic TRAX monomers at different positions within the translin octamer might modify the equilibrium population of conformers and hence the binding affinity for nucleic acids and the catalytic activity, perhaps tuning the dynamics and the biological activity of the assembly while keeping the overall architecture.
Conformational heterogeneity of nucleic acids binding to translin
Translin has been shown to bind ssRNA and ssDNA in solution (2,7–9,29), which has been linked to possible roles in mRNA translation and transport, RNAi metabolism (2,12,15,17,18,51,52) and tRNA processing (19), as well as in other possible functions of DNA metabolism (7–9). Remarkably, while the previously identified nucleic acid-binding region (28) is not accessible for nucleic acids binding in the compact (crystallographic) structure, it becomes accessible in the open configurations of translin in solution found here based on SAXS data (Figure 1b). This suggests that the binding of translin to nucleic acids is related to the conformational equilibrium of the protein in solution, in which the open conformations could facilitate the process of recognition. Interestingly, the dimensions of the groove in the open conformations (length ∼60 Å; width ∼25 Å) would be too tight for accommodating a double-stranded DNA molecule, in agreement with the experimentally observed inability of translin to bind double-stranded DNA (8). According to the similarity of the p(r) functions derived from the free and bound states (Supplementary Figure S1), the presence of ssRNA does not rigidify translin. The repetitive sequence of the intrinsically flexible ssRNA (GU)12 that hosts several binding regions to translin, and the presence of multiple and symmetric binding sites within translin cavity produces a multiplicity of potential low-affinity interactions that do not perturb the inherent breathing of translin. This observation is coherent with the large conformational variability of ssRNA that was found to be compatible with the SAXS data. Based on these considerations, we speculate that the translin octamer conformational dynamics might be essential for the accommodation of different RNA or DNA molecules, and such model could provide an explanation for the differential affinities of the protein for specific RNA and DNA sequences (8,29).
Implications for the biological functions of translin and TRAX proteins
Drosophila and human translin–TRAX complexes were found to have an endonucleolytic activity, which degrades the ago2-nicked passenger RNA strand in RNA-induced silencing complexes (RISC), thereby activating those RISC complexes (15,17,18). In Neurospora such complexes appear to be involved in the removal of 5′ pre-tRNA fragments after the processing of pre-tRNAs by RNase P (19). The catalytic sites for these degradation activities reside in the TRAX subunits. Although the structural studies reported in this article were all carried out with pure recombinant translin complexes and the presence of TRAX could introduce subtle changes in the dynamics of the translin–TRAX octamers (as discussed above), it appears reasonable to think that the overall transitions will be the same, and it would be interesting to explore in the future whether such transitions could have a role in the endonuclease activity of the system. It should also be noted that pure translin octamers can be formed in cells in addition to translin–TRAX complexes. Translin octamers could be engaged in other functions that have been previously suggested, such as participation in specific mRNA trafficking along dendrites of nerve cells (52). In fact, it appears that activities such as RNA transport might be carried out by pure translin octamers, given that they can bind RNA, but lack the endonuclease activity of TRAX. In this way, the cargo RNA will be maintained intact during the translocation activity. Furthermore, the localization of RNA in the internal cavity of pure translin complexes (as shown in this article), could protect the transported mRNA from being translated in the inappropriate location, or from being degraded by other nucleases, which would help to maintain its functionality (51). Interestingly, only TRAX has a nuclear localization signal, while translin has just nuclear export signal (4,5,53). Thus, pure translin octamers would be expected to remain in the cytosplasm and carry out cytoplasmic functions such as mRNA trafficking in dendrites. In summary, the inherent plasticity of translin assembly shown in this work provides a structural basis for its diverse biological roles.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [54–58].
FUNDING
FPU fellowship from the Spanish Ministry of Science (to L.P.-C.); fellowship from the Clore Foundation PhD Scholars program (to K.L.); Spanish Plan Nacional I+D+i [BIO2010-22324]; SPIN-HD—ANR Chaires d’Excellence from the Agence National de Recherche; Israel Science Foundation [967/08]. Funding for open access charge: Spanish Plan Nacional I+D+i (BIO2010-22324) and the SPIN-HD—ANR Chaires d’Excellence from the Agence National de Recherche.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Assistance at the X33 EMBL beamline is acknowledged. We thank Dr M. Kasai of the Department of Immunology, National Institutes of Health, Tokyo, Japan, for sending us the plasmid encoding the human translin. Emmanuel Margeat (CBS-Montpellier) and Manuel Rueda (BSC) are acknowledged for useful discussions.
REFERENCES
- 1.Aharoni A, Baran N, Manor H. Characterization of a multisubunit human protein which selectively binds single stranded d(GA)n and d(GT)n sequence repeats in DNA. Nucleic Acids Res. 1993;21:5221–5228. doi: 10.1093/nar/21.22.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han JR, Gu W, Hecht NB. Testis-brain RNA-binding protein, a testicular translational regulatory RNA-binding protein, is present in the brain and binds to the 3' untranslated regions of transported brain mRNAs. Biol. Reprod. 1995;53:707–717. doi: 10.1095/biolreprod53.3.707. [DOI] [PubMed] [Google Scholar]
- 3.Kasai M, Aoki K, Matsuo Y, Minowada J, Maziarz RT, Strominger JL. Recombination hotspot associated factors specifically recognize novel target sequences at the site of interchromosomal rearrangements in T-ALL patients with t(8;14)(q24;q11) and t(1;14)(p32;q11) Int. Immunol. 1994;6:1017–1025. doi: 10.1093/intimm/6.7.1017. [DOI] [PubMed] [Google Scholar]
- 4.Aoki K, Ishida R, Kasai M. Isolation and characterization of a cDNA encoding a Translin-like protein, TRAX. FEBS Lett. 1997;401:109–112. doi: 10.1016/s0014-5793(96)01444-5. [DOI] [PubMed] [Google Scholar]
- 5.Chennathukuzhi VM, Kurihara Y, Bray JD, Hecht NB. Trax (translin-associated factor X), a primarily cytoplasmic protein, inhibits the binding of TB-RBP (translin) to RNA. J. Biol. Chem. 2001;276:13256–13263. doi: 10.1074/jbc.M009707200. [DOI] [PubMed] [Google Scholar]
- 6.Gupta GD, Kale A, Kumar V. Molecular evolution of translin superfamily proteins within the genomes of eubacteria, archaea and eukaryotes. J. Mol. Evol. 2012;75:155–167. doi: 10.1007/s00239-012-9534-z. [DOI] [PubMed] [Google Scholar]
- 7.Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat. Genet. 1995;10:167–174. doi: 10.1038/ng0695-167. [DOI] [PubMed] [Google Scholar]
- 8.Jacob E, Pucshansky L, Zeruya E, Baran N, Manor H. The human protein translin specifically binds single-stranded microsatellite repeats, d(GT)n, and G-strand telomeric repeats, d(TTAGGG)n: a study of the binding parameters. J. Mol. Biol. 2004;344:939–950. doi: 10.1016/j.jmb.2004.09.095. [DOI] [PubMed] [Google Scholar]
- 9.Kaluzhny D, Laufman O, Timofeev E, Borisova O, Manor H, Shchyolkina A. Conformational changes induced in the human protein translin and in the single-stranded oligodeoxynucleotides d(GT)(12) and d(TTAGGG)(5) upon binding of these oligodeoxynucleotides by translin. J. Biomol. Struct. Dyn. 2005;23:257–265. doi: 10.1080/07391102.2005.10507064. [DOI] [PubMed] [Google Scholar]
- 10.Chennathukuzhi VM, Kurihara Y, Bray JD, Yang J, Hecht NB. Altering the GTP binding site of the DNA/RNA-binding protein, Translin/TB-RBP, decreases RNA binding and may create a dominant negative phenotype. Nucleic Acids Res. 2001;29:4433–4440. doi: 10.1093/nar/29.21.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkenstadt PM, Kang WS, Jeon M, Taira E, Tang W, Baraban JM. Somatodendritic localization of Translin, a component of the Translin/Trax RNA binding complex. J. Neurochem. 2000;75:1754–1762. doi: 10.1046/j.1471-4159.2000.0751754.x. [DOI] [PubMed] [Google Scholar]
- 12.Morales CR, Lefrancois S, Chennathukuzhi V, El-Alfy M, Wu X, Yang J, Gerton GL, Hecht NB. A TB-RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev. Biol. 2002;246:480–494. doi: 10.1006/dbio.2002.0679. [DOI] [PubMed] [Google Scholar]
- 13.Wu XQ, Hecht NB. Mouse testis brain ribonucleic acid-binding protein/translin colocalizes with microtubules and is immunoprecipitated with messenger ribonucleic acids encoding myelin basic protein, alpha calmodulin kinase II, and protamines 1 and 2. Biol. Reprod. 2000;62:720–725. doi: 10.1095/biolreprod62.3.720. [DOI] [PubMed] [Google Scholar]
- 14.Yang S, Cho YS, Chennathukuzhi VM, Underkoffler LA, Loomes K, Hecht NB. Translin-associated factor X is post-transcriptionally regulated by its partner protein TB-RBP, and both are essential for normal cell proliferation. J. Biol. Chem. 2004;279:12605–12614. doi: 10.1074/jbc.M313133200. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Boja ES, Oubrahim H, Chock PB. Testis brain ribonucleic acid-binding protein/translin possesses both single-stranded and double-stranded ribonuclease activities. Biochemistry. 2004;43:13424–13431. doi: 10.1021/bi048847l. [DOI] [PubMed] [Google Scholar]
- 17.Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y, Simanshu DK, Ascano M, Diaz-Avalos R, Park AY, Juranek SA, Rice WJ, Yin Q, Robinson CV, Tuschl T, et al. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat. Struct. Biol. 2011;18:658–664. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Gu W, Liang C, Liu Q, Mello CC, Liu Y. The translin-TRAX complex (C3PO) is a ribonuclease in tRNA processing. Nat. Struct. Mol. Biol. 2012;19:824–830. doi: 10.1038/nsmb.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu XQ, Xu L, Hecht NB. Dimerization of the testis brain RNA-binding protein (translin) is mediated through its C-terminus and is required for DNA- and RNA-binding. Nucleic Acids Res. 1998;26:1675–1680. doi: 10.1093/nar/26.7.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasai M, Matsuzaki T, Katayanagi K, Omori A, Maziarz RT, Strominger JL, Aoki K, Suzuki K. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J. Biol. Chem. 1997;272:11402–11407. doi: 10.1074/jbc.272.17.11402. [DOI] [PubMed] [Google Scholar]
- 22.VanLoock MS, Yu X, Kasai M, Egelman EH. Electron microscopic studies of the translin octameric ring. J. Struct. Biol. 2001;135:58–66. doi: 10.1006/jsbi.2001.4383. [DOI] [PubMed] [Google Scholar]
- 23.Yu E, Owttrim GW. Characterization of the cold stress-induced cyanobacterial DEAD-box protein CrhC as an RNA helicase. Nucleic Acids Res. 2000;28:3926–3934. doi: 10.1093/nar/28.20.3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X, Hingorani MM, Patel SS, Egelman EH. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat. Struct. Biol. 1996;3:740–743. doi: 10.1038/nsb0996-740. [DOI] [PubMed] [Google Scholar]
- 25.Pascal JM, Hart PJ, Hecht NB, Robertus JD. Crystal structure of TB-RBP, a novel RNA-binding and regulating protein. J. Mol. Biol. 2002;319:1049–1057. doi: 10.1016/S0022-2836(02)00364-9. [DOI] [PubMed] [Google Scholar]
- 26.Sugiura I, Sasaki C, Hasegawa T, Kohno T, Sugio S, Moriyama H, Kasai M, Matsuzaki T. Structure of human translin at 2.2 A resolution. Acta. Crystallogr. D. Biol. Crystallogr. 2004;60:674–679. doi: 10.1107/S0907444904002549. [DOI] [PubMed] [Google Scholar]
- 27.Kumar V, Gupta GD. Low-resolution structure of Drophila translin. FEBS Open Biol. 2012;2:37–46. doi: 10.1016/j.fob.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eliahoo E, Ben Yosef R, Perez-Cano L, Fernandez-Recio J, Glaser F, Manor H. Mapping of interaction sites of the Schizosaccharomyces pombe protein Translin with nucleic acids and proteins: a combined molecular genetics and bioinformatics study. Nucleic Acids Res. 2010;38:2975–2989. doi: 10.1093/nar/gkp1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laufman O, Ben Yosef R, Adir N, Manor H. Cloning and characterization of the Schizosaccharomyces pombe homologs of the human protein Translin and the Translin-associated protein TRAX. Nucleic Acids Res. 2005;33:4128–4139. doi: 10.1093/nar/gki727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin RG, Ames BN. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J. Biol. Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- 31.Konarev PV, Volkov VV, Sokolova AV, Koch MH, Svergun DI. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 32.Fischer H, Oliveira MM, Napolitano HB, Craievich AF, Polikarpov I. The molecular weight of proteins in solution can be determined from a single SAXS measurement on a relative scale. J. Appl. Cryst. 2010;43:101–109. [Google Scholar]
- 33.Guinier A. La diffraction des rayons X aux tres petits angles; application a l'etude de phenomenes ultramicroscopiques. Ann. Phys. 1939;12:161–237. [Google Scholar]
- 34.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 35.Svergun DI, Barberato C, Koch MH. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 36.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 37.Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emperador A, Carrillo O, Rueda M, Orozco M. Exploring the suitability of coarse-grained techniques for the representation of protein dynamics. Biophys. J. 2008;95:2127–2138. doi: 10.1529/biophysj.107.119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rueda M, Chacon P, Orozco M. Thorough validation of protein normal mode analysis: a comparative study with essential dynamics. Structure. 2007;15:565–575. doi: 10.1016/j.str.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Lopez-Blanco JR, Garzon JI, Chacon P. iMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27:2843–2850. doi: 10.1093/bioinformatics/btr497. [DOI] [PubMed] [Google Scholar]
- 41.Abagyan R, Totrov MM, Kuznetsov DA. ICM: a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994;15:488–506. [Google Scholar]
- 42.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 43.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 44.Flores SC, Sherman MA, Bruns CM, Eastman P, Altman RB. Fast flexible modeling of RNA structure using internal coordinates. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011;8:1247–1257. doi: 10.1109/TCBB.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koch MH, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q. Rev. Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 47.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 48.Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Cano L, Fernandez-Recio J. Optimal protein-RNA area, OPRA: a propensity-based method to identify RNA-binding sites on proteins. Proteins. 2010;78:25–35. doi: 10.1002/prot.22527. [DOI] [PubMed] [Google Scholar]
- 50.Tama F, Gadea FX, Marques O, Sanejouand YH. Building-block approach for determining low-frequency normal modes of macromolecules. Proteins. 2000;41:1–7. doi: 10.1002/1097-0134(20001001)41:1<1::aid-prot10>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 51.Han JR, Yiu GK, Hecht NB. Testis/brain RNA-binding protein attaches translationally repressed and transported mRNAs to microtubules. Proc. Natl Acad. Sci. USA. 1995;92:9550–9554. doi: 10.1073/pnas.92.21.9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc. Natl Acad. Sci. USA. 2009;106:16481–16486. doi: 10.1073/pnas.0902833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho YS, Chennathukuzhi VM, Handel MA, Eppig J, Hecht NB. The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J. Biol. Chem. 2004;279:31514–31523. doi: 10.1074/jbc.M401442200. [DOI] [PubMed] [Google Scholar]
- 54.Lasker K, Topf M, Sali A, Wolfson HJ. Inferential optimization for simultaneous fitting of multiple components into a CryoEM map of their assembly. J. Mol. Biol. 2009;388:180–194. doi: 10.1016/j.jmb.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng TM, Blundell TL, Fernandez-Recio J. pyDock: electrostatics and desolvation for effective scoring of rigid-body protein-protein docking. Proteins. 2007;68:503–515. doi: 10.1002/prot.21419. [DOI] [PubMed] [Google Scholar]
- 56.Fernández-Recio J, Totrov M, Skorodumov C, Abagyan R. Optimal Docking Area: a new method for predicting protein-protein interaction sites. Proteins. 2005;58:134–143. doi: 10.1002/prot.20285. [DOI] [PubMed] [Google Scholar]
- 57.Aoki K, Suzuki K, Ishida R, Kasai M. The DNA binding activity of Translin is mediated by a basic region in the ring-shaped structure conserved in evolution. FEBS Lett. 1999;443:363–366. doi: 10.1016/s0014-5793(99)00010-1. [DOI] [PubMed] [Google Scholar]
- 58.Mashiach-Farkash E, Nussinov R, Wolfson HJ. SymmRef: a flexible refinement method for symmetric multimers. Proteins. 2011;79:2607–2623. doi: 10.1002/prot.23082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.