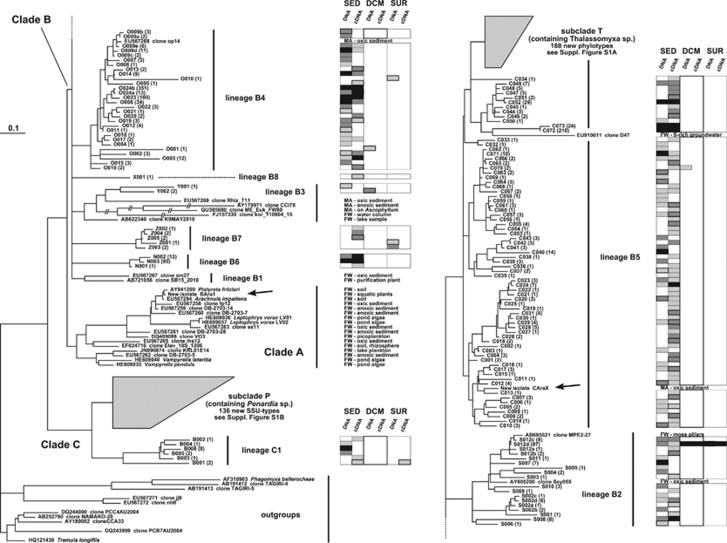
Figure 5.
Graphical representation of the diversity of vampyrellid SSU-types found in the BioMarKs data in relation to sequenced isolates, highlighting their habitat preferences. A ML tree is used, based on the V4 region of the SSU rDNA; it was rooted with selected members of other endomyxan lineages. Clades and lineages are labelled consistently with Figure 4. Some branch lengths within fast-evolving lineage B3 have been reduced by half. Sequences from our new isolates are highlighted with arrows. The ecological provenance of environmental sequences from GenBank is listed (see also Supplementary Table S2). That of BioMarKs V4 SSU-types is indicated in the boxes (SED, sediment; DCM, deep chlorophyll maximum; SUR, surface waters). In the labels of the BioMarKs SSU-types, the number in brackets indicates how many individual sequences belong to that SSU-type. For each SSU-type, varying shades of grey in the boxes give an indication of the number of individual sequences found in each particular sample type: light grey for unique sequences, medium grey for 2 to 5 sequences, dark grey for 6 to 20 sequences, and black for >20 sequences. The two most diverse lineages (subclades P and T) were collapsed for clarity, but the V4 diversity and ecological provenance of all SSU-types within these lineages is provided in Supplementary Figure S1.