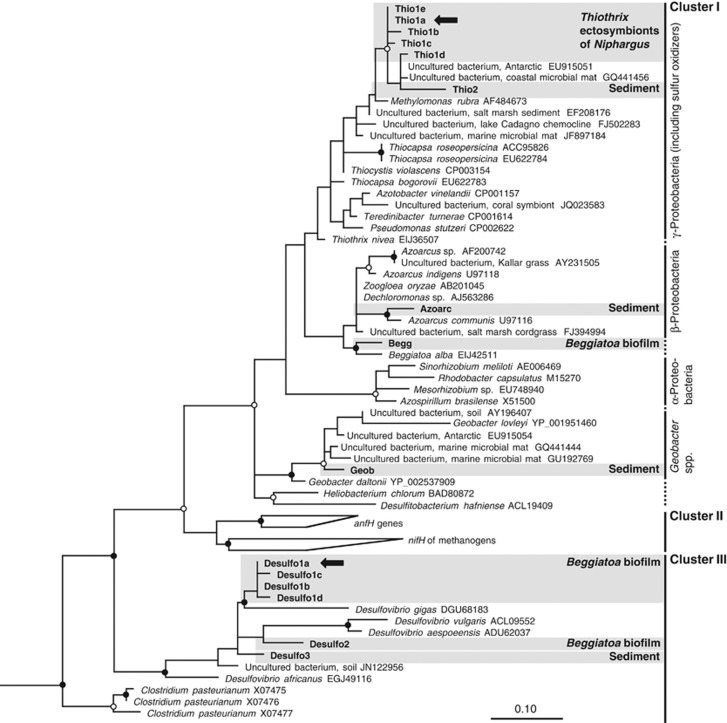
Figure 2.
Maximum-likelihood tree exhibiting the phylogenetic relationship of nifH transcripts and genes derived from Niphargus spp., Beggiatoa-dominated biofilms and sulfidic sediment within the Frasassi caves with closely related and prototypical isolates and sequences obtained from other habitats. One hundred and three unambiguously aligned amino-acid positions were considered. Sequences from this study are shown in bold; when numerous closely related phylotypes were obtained from a particular sample, the preferentially expressed phylotype is shown with arrows (see Table 2 for details). Cluster nomenclature is based on Zehr et al. (2003b); vertical lines denote distinct nifH Clusters, and for Cluster I, different groups are connected with dotted lines. The tree was rooted using nifH gene of Methanosarcina barkeri (GenBank accession no. AB019139), which falls in Cluster IV. Tree topology was validated by maximum-parsimony analysis; nodes with strong bootstrap values are marked (○>50% ●>70%). For tentative assignment of different phylotypes to specific bacterial groups, see Discussion.