Figure 2.
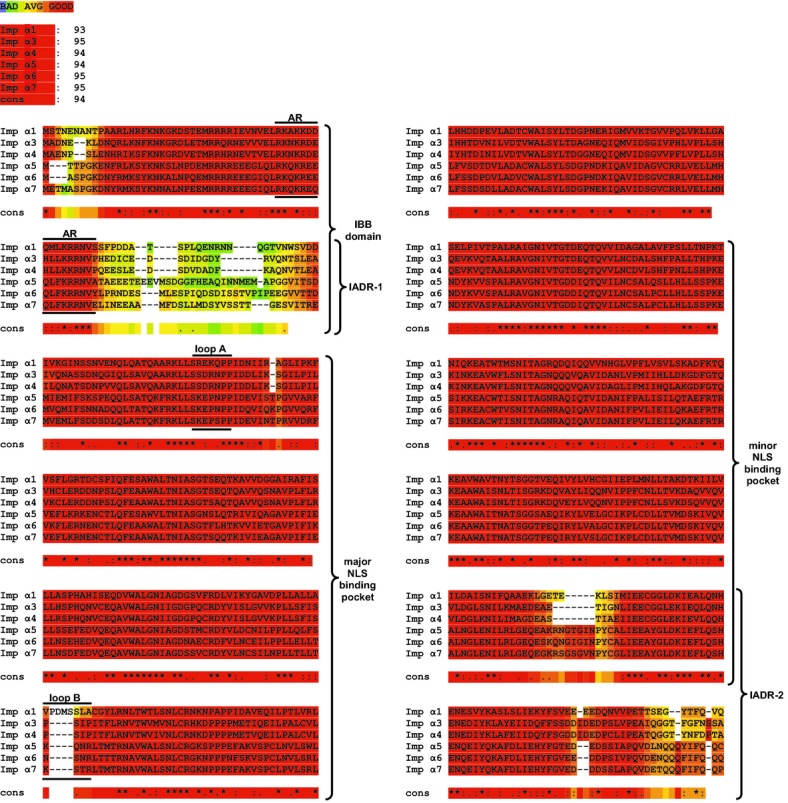
Sequence alignment. Multiple sequence alignment of the importin α family proteins by T‐Coffee software, displayed as a “heat map.” Regions with a high degree of similarity are shown in red, with decreasing similarity indicated from orange to blue (see color key at top). The overall similarity between different proteins is shown immediately below the color key, followed by the individual sequence alignments. The graphic colored output indicates the level of consistency between the final alignment and the library used by T‐Coffee. The main score is the total consistency value. A value of a 100 means full agreement between the considered alignment and its associated primary library. Please note the 2 regions of sequence divergence, denoted importin α diversity regions (IADRs), positioned near the beginning and end of the sequences. IADR‐1 and IADR‐2 are adjacent to, or overlap, the major and minor nuclear localization sequence (NLS) binding pockets, respectively. Also indicated are the positions of the auto‐inhibitory region (AR) within the importin β binding (IBB) domain, and loops A and B within the major NLS binding pocket. Symbol key: “cons” represents the consensus sequence, “*” represents the same amino acid at a given position in all proteins, “:” represents highly similar residues at a given position, and “.” represents amino acids with a similar functionality at a given position.
