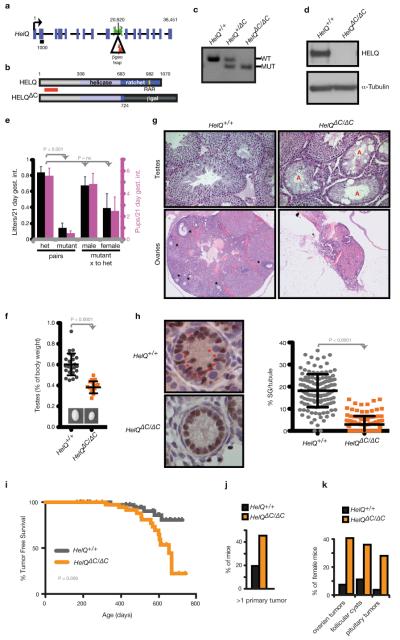
Figure 1. A mouse model of HELQ deficiency.
a, HelQ genomic locus; base pairs are indicated above; introns are not to scale; exons are roughly to scale with black bar indicating 1 kb; location of the β-geo trap and genotyping primers are shown. b, HELQ domain architecture: amino acids are indicated, red bar spans epitope recognized by the HELQ antibody used for western blotting. c, HelQ genotype PCR. d, Lysates from ear fibroblasts of HelQ mice probed for HELQ. e, The number of litters (black) and pups (pink) generated per 21-day gestational interval. f, HelQ testis images and weights. g, Histological sections of HelQ gonads. Atrophic tubules (A); developing ovarian follicles (*). h, 5-day-old neonatal testes labelled with the Sertoli marker WT1 (brown) and hematoxylin (blue), to reveal spermatogonia (*); quantification of spermatogonia (SG). i, Epithelial and stromal tumour free survival of HelQ mice. j, Frequency of mice with 2 or more primary tumours, and k, female-specific pathology.