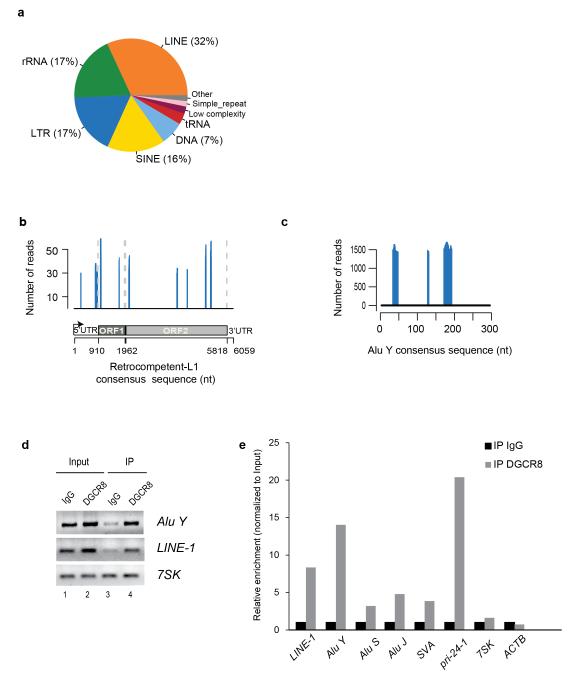
Figure 1.
DGCR8 binds a constellation of transcripts from repetitive elements. (a) Pie chart showing distribution of reads mapping to repetitive elements in sense and antisense orientation. 73% of the significant clusters map to transposable elements including DNA-Transposons, LINE-1s, LTR-containing retrotransposons and SINEs. (b) Distribution of DGCR8 binding sites in a human RC-L1Hs consensus sequence (FDR <0.01). Only sense peaks with a minimum of 29 reads are represented. Schematic representation of the RC-L1 element (bottom). UTR, untranslated region; ORF, open reading frame. (c) Distribution of significant DGCR8 binding sites in a human Alu Y consensus sequence in sense orientation (FDR <0.01). (d) Amplification of Alu Y, LINE-1 and 7SK mRNAs by RT-PCR upon immunoprecipitation of endogenous DGCR8 protein. (e) Real-time Reverse Transcriptase (RT)-PCR analysis of several transposable elements (LINE-1, Alu Y, Alu S Alu J and SVA) upon immunoprecipitation of endogenous DGCR8 of one representative experiment. Grey bars represent the relative enrichment over control IgG immunoprecipitation (black bars) and data is expressed as a percentage of input for normalization purposes. In this assay, a well-known target of the Microprocessor (pri-miR-24-1) was used as an internal positive control, whereas 7SK and ACTB were used as negative controls. IP,immunoprecipitation.