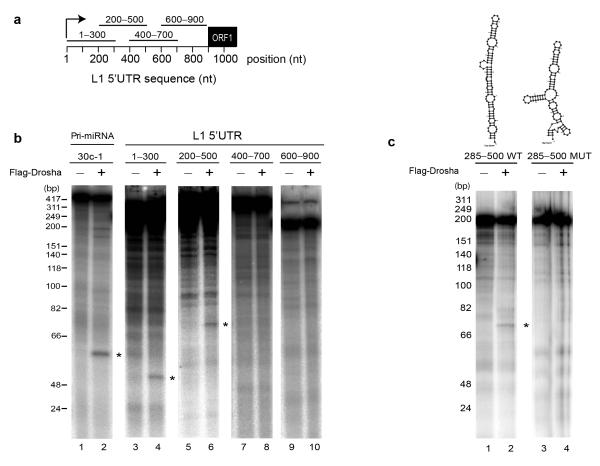
Figure 3.
The 5′UTR of L1 mRNA is cleaved by immunopurified Drosha in vitro. (a) Schematic representation of four 300-nucleotide in vitro transcribed fragments spanning the sense L1 5′UTR region used in (b) for in vitro processing. Transcripts were incubated (+) or not (−) with immunopurified Microprocessor (using FLAG-Drosha). Cleavage products are indicated with asterisks (lanes 2, 4, 6). An RNA ladder marker indicates sizes in base-pairs on the left. (c) In vitro processing of 285-500 L1 5′UTR region (lane 2). Top panel shows a predicted pri-miRNA-like structure of this region. Cleavage is abolished upon introducing mutations that disturb this structure (lane 4 and top panel).