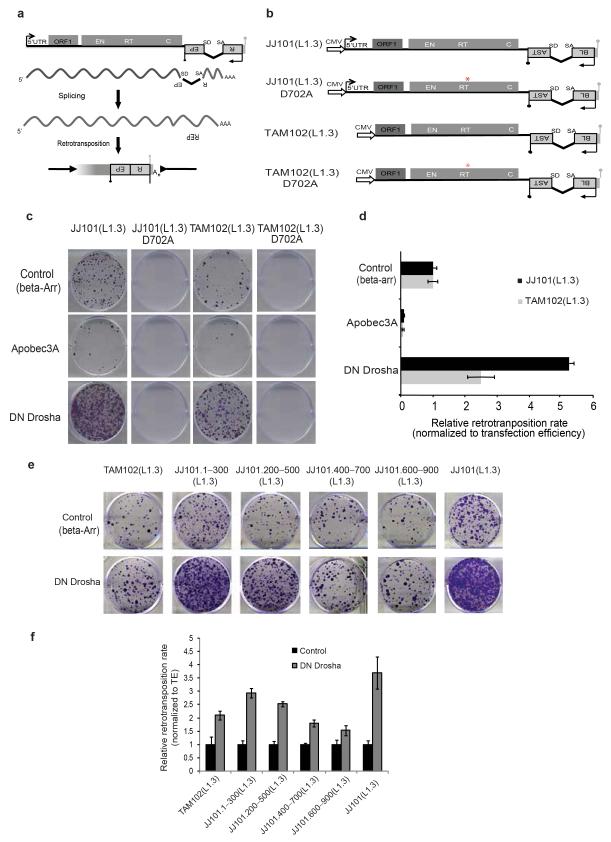
Figure 4.
The Microprocessor negatively regulates L1 retrotranspositionin vivo. (a) Cartoon depicting the LINE-1 based retrotransposition assay in cultured cells. The transcription start site at the L1 5′ UTR (black arrow), the L1 open reading frames (ORF1 and ORF2; gray rectangles), and the L1 poly (A) site (grey lollipop) are indicated. The relative locations of the endonuclease (EN), reverse transcriptase (RT), and cysteine-rich (C) domains of ORF2 are also indicated. (b) Schematic representation of the JJ101(L1.3) vector containing a full-length human RC-L1 sequence tagged with a mblastI cassette (Blasticidin-resistance gene). The 5′UTR of L1.3 was removed to generate construct TAM102(L1.3). Alleles containing a missense mutation in the Reverse Transcriptase (RT) domain of L1-ORF2 (red asterisk, JJ101 (L1.3) D702A and TAM102 (L1.3) D702A) were used as an internal control. CMV denotes the presence of a Cytomegalovirus promoter. Black and grey lollipops represent polyadenylation signals. (c,e) Cell culture-based LINE-1 retrotranposition assay. (c) HeLa cells were co-transfected with JJ101(L1.3), TAM102(L1.3) and corresponding control vectors or the indicated engineered LINE-1 construct containing an individual fragment derived from the 5′UTR region plus the indicated expression plasmids (β-arrestin (β-arr), Apobec3A, or DN Drosha), as indicated on the left. Each image shows representative data from L1 retrotransposition assays conducted in triplicate. (d,f) Quantification of LINE-1 retrotranposition (from panel c and e respectively). Blasticidin-resistant foci were manually counted and quantified. Data is presented as the proportion of the activity seen in cultures co-transfected with the plasmid expressing the negative control (β-arrestin) and normalized using transfection efficiency and toxicity. Shown are averages of 3 independent biological replicates ± s.d.