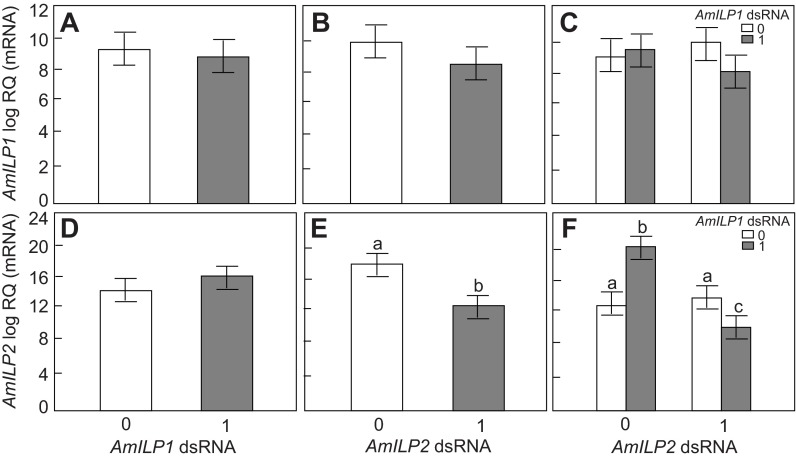
Fig. 1.
Apis mellifera insulin-like peptide AmILP1 and AmILP2 gene knockdown validation in fifth instar honey bee larvae. (A,B,D,E) The main effects of AmILP1 double-stranded (ds)RNA and AmILP2 dsRNA on AmILP1 and AmILP2 gene expression in a factorial ANOVA. (C,F) The relationships between four treatment groups, as revealed by a Fisher's least significant difference (LSD) post hoc test: the bars from left to right represent gfp (green fluorescent protein), AmILP1 dsRNA, AmILP2 dsRNA, and AmILP1 dsRNA plus AmILP2 dsRNA. Overall, AmILP1 gene expression was not affected by AmILP1 dsRNA (A) and AmILP2 dsRNA (B). Overall, AmILP2 gene expression was not affected by AmILP1 dsRNA (D), but was significantly reduced by AmILP2 dsRNA (E). Panel F shows that compared with gfp (first bar): (i) AmILP1 dsRNA treatment increased AmILP2 expression level; (ii) AmILP2 dsRNA in the absence of AmILP1 dsRNA did not reduce the AmILP2 mRNA level; (iii) but when combined with AmILP1 dsRNA application, AmILP2 was significantly downregulated. Data are presented as means ± s.e.m. (N=15). RQ is relative quantification. Different letters (a–c) over the bars indicate significant differences between treatments. ‘0’ represents no dsRNA treatment and ‘1’ represents dsRNA treatment.