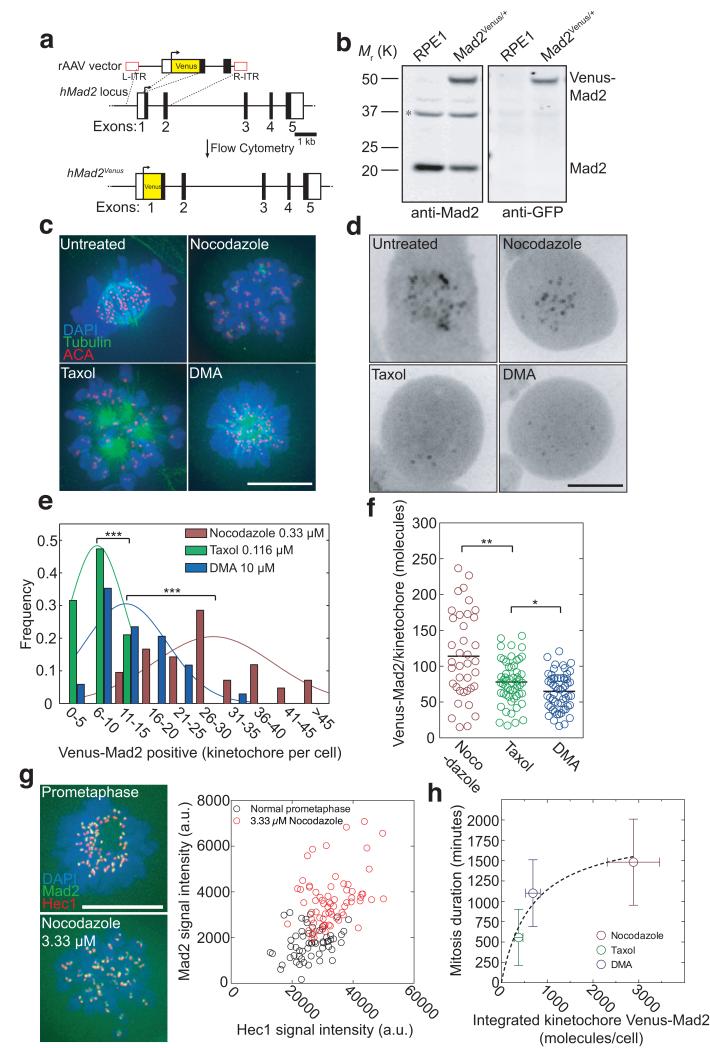
Fig. 3. The amount of Mad2-positive kinetochores correlates with the length of a mitotic block.
(a) Venus ORF targeting into the hMAD2L1 locus by rAAV-mediated homologous recombination. The rAAV vector contained the coding sequence of Venus, which was inserted between two gene-specific homology arms to replace the ATG at the junction between non-coding (white square) and coding (black square) regions of exon 1 and two inverted terminal repeats (L-ITR, R-ITR; red rectangles). Integrants were selected by FACS. (b) Western blot analysis of parental RPE1 and a Venus-Mad2 clone. Cell lysates were probed with anti-Mad2 (left panel) and anti-GFP (right panel) antibodies. The asterisk indicates a cross-reacting band. Note the two forms of Mad2 in the Venus-Mad2 clone (the upper one uniquely recognized by the anti-GFP antibody). Molecular mass markers on the left. (c) Asynchronous RPE1 cells were treated for 12-16 hours with nocodazole, or Taxol or DMA, fixed with methanol and processed for immunofluorescence using anti-beta-tubulin (green) and ACA antibodies to stain the kinetochores (red). DNA was stained using DAPI (blue). Merged images are presented. Representative of 20-50 cells from at least two independent experiments. (d) Typical localization of Venus-Mad2 in living mitotic cells treated with different spindle poisons as in c). (e) Distribution of Venus-Mad2-positive kinetochores per cell following 12-16 hours of treatment with different drugs (n=42 cells for nocodazole; n=38 cells for Taxol and n=34 cells for DMA), representative of at least two independent experiments. (f) Scatter plots obtained by fluorescence correlation using data shown in Supplementary Fig. 4 of the number of Venus-Mad2 molecules per kinetochore in cells treated with different drugs (n=57 kinetochores from 8 cells for nocodazole; n=38 kinetochores from 11 cells for Taxol and n=55 kinetochores from 8 cells for DMA), representative of at least two independent experiments. The black line represents the mean. (g) Asynchronous (top) or 3.33 μM nocodazole-treated (bottom) RPE1 Venus-Mad2 cells were fixed in methanol and processed for immunofluorescence against Hec1 (red), and DNA stained with DAPI (blue). The signal intensity for Hec1, a stable kinetochore component, was used as a measure of kinetochore size. Kinetochore-associated Mad2 signals (green in image) were quantified and plotted as a function of the corresponding Hec1 fluorescence (n=56 kinetochores from 6 cells for asynchronous and n=60 kinetochores from 3 cells for 3.33 μM nocodazole). Data are representative of two independent experiments. (h) Plot of the relationship between the duration of mitosis (from Fig. 2c) and the integrated molecule number of Venus-Mad2 at kinetochores in the different poisons (from Fig. 3e,f). Error bars indicate s.d. and are representative of at least two independent experiments. Scale bars = 10 μm.