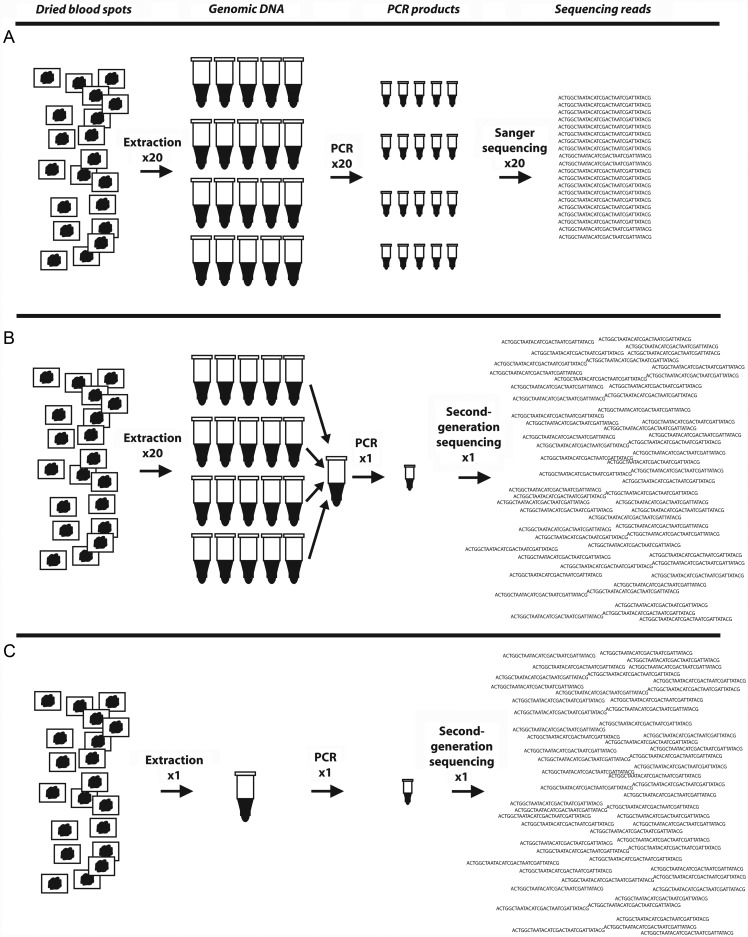
Figure 1.
Schematic of different sequencing approaches used to characterize prevailing parasite genotypes. A, Specimens are collected from patients, genomic DNA (gDNA) is extracted individually, target sequences are amplified individually, and polymerase chain reaction (PCR) products are sequenced individually to obtain a sequencing read for each isolate. These reads are then used to generate a prevalence of mutant alleles or combined with an estimate of each isolate's multiplicity of infection to estimate the frequency of alleles. B, Specimens are collected from patients, gDNA is extracted individually but then combined into a gDNA pool, the target sequence is amplified from this pooled gDNA using primers adapted for second-generation sequencing (eg, 454, Illumina), and this PCR product is sequenced on a second-generation sequencing platform to yield thousands of sequencing reads. These reads are then aligned to a reference sequence and scored for mutations to estimate the frequency of mutant alleles. C, Specimens are collected from patients, but gDNA is extracted in a single pool, and the procedure follows as described for B.