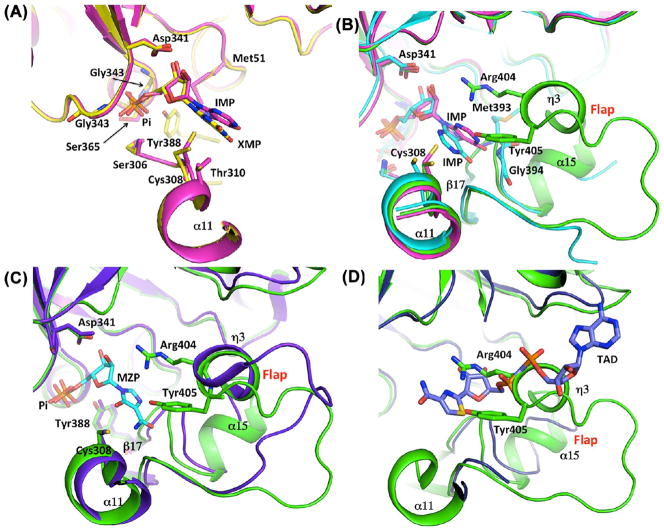
Figure 4.
Structural alignment of IMPDH active sites. (A) Overlap of the B. anthracis IMPDH structures of Pi (green), IMP (magenta), and XMP (yellow) complexes showing the positions of the IMP and XMP (magenta and yellow sticks, respectively). (B) Overlap of the B. anthracis Pi-bound (green) and IMP-bound (magenta) structures with the S. pyogenes enzyme-IMP complex (PDB entry 1ZFJ) (turquoise). IMP molecules (B. anthracis in magenta, S. pyogenes in turquoise) are depicted as sticks. S. pyogenes residues Met393 and Gly394 interacting with IMP are also depicted as sticks. (C) Overlap of the structures of the B. anthracis Pi-bound enzyme (green) with the T. foetus IMPDH complex with MZP (purple) (PDB entry 1PVN20). The conserved Arg-Tyr dyad within helix η 3 of the flap is depicted as sticks. (D) Overlap of the structures of the B. anthracis Pi-bound enzyme (green) with the T. foetus IMPDH (purple) complex with IMP (omitted for the sake of clarity) and TAD (purple) (PDB entry 1LRT19). A distal portion of the B. anthracis catalytic flap with the conserved Arg-Tyr dyad is clashing with the T. foetus β-methylene-thiazole portion of TAD, indicating that these two elements are occupying the same space within the active site.