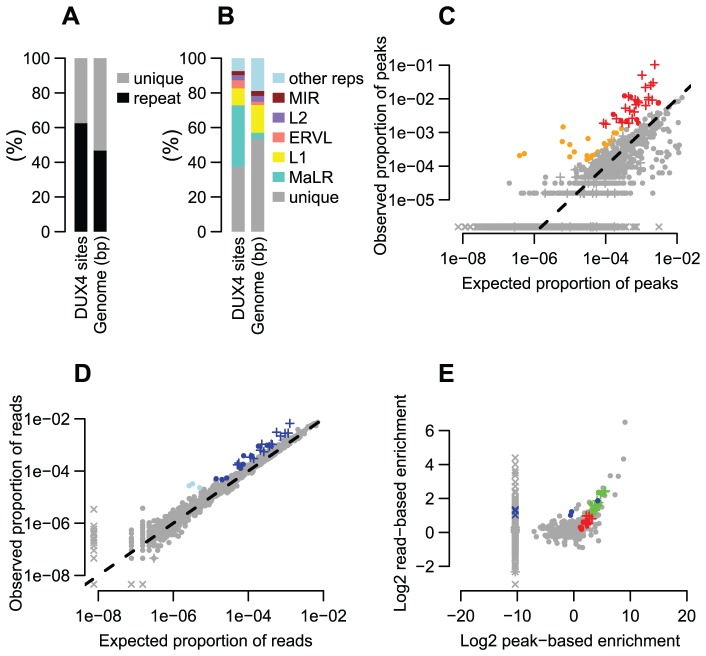
Figure 1. Many repeat types are enriched among DUX4 binding sites.
(A) ∼2/3 of DUX4 binding-sites are in repetitive elements, compared to ∼45% of the human genome. (B) Comparing repeat family proportions among DUX4 binding-sites with genome-wide fractions shows ∼10-fold MaLR enrichment. (C) A peak-based method of estimating repeat enrichment uses uniquely-mapped reads, so is blind to recently active repeats; however, it ignores background reads so provides a more sensitive enrichment measure than the read-based estimate (Figures 1D, 1E). 32 repeat types (red) are enriched ≥2-fold with ≥100 peaks (arbitrary thresholds) (Tables 1, S2 and S3); 21 (orange) are rarer in the genome (10–99 peaks) but enriched ≥4-fold. The log10-scaled x-axis shows the proportion of peaks expected to overlap each repeat type if DUX4 binding sites had uniform genomic distribution; the log10-scaled y-axis shows observed proportions. The dashed line represents no enrichment. In all panels, “+” symbols represent MaLR elements and “x” datapoints represent repeat types for which no peaks/reads were observed – these are given an arbitrary low (non-zero) value to ensure visibility on log-scaled plots. (D) The read-based enrichment estimation method examines highly similar repeats as well as uniquely-mappable sequences, but gives a “dampened” enrichment measure due to background reads in ChIP-seq samples (see Methods). 25 repeat types (dark blue) are enriched ≥2-fold, with ≥1000 reads (arbitrary thresholds); 3 (light blue) are rarer among ChIP-seq reads (100–999 reads) but enriched ≥4-fold. (E) The peak-based (x-axis) and read-based (y-axis) methods yield similar results. 19 repeat types (green datapoints, thresholds as in Figures 1A and B for red and dark blue points) were enriched in both analyses; 13 enriched only by the peak-based method (red), and 6 enriched only by the read-based method (blue). Additional repeats (gray datapoints, upper-right quadrant) appear enriched by both methods, but are rare in the genome so do not exceed our arbitrary peak/read thresholds.