Figure 6. Read-based enrichment analysis of RNA-seq data shows activation of several repeat types.
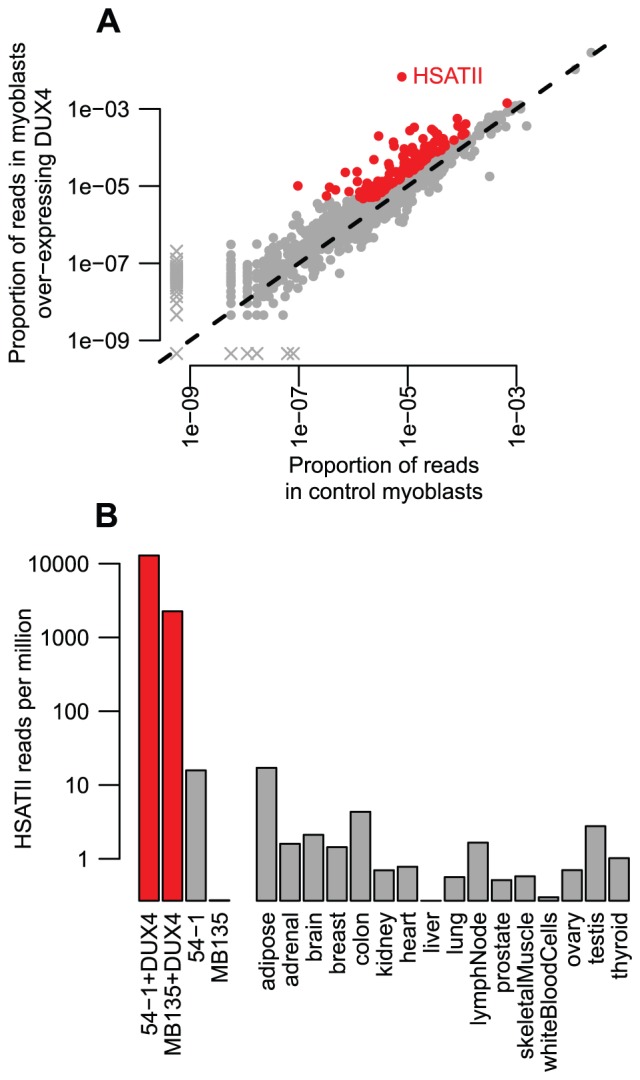
(A) For each repeat type, we show the proportion of RNA-seq reads in two myoblast cell lines transduced with lentiviral DUX4 (y-axis) compared to the proportion from the same two myoblast cell lines without DUX4 transduction (x-axis). Repeats plotted in red show ≥2-fold activation and ≥1000 reads summed across the two DUX4-expressing myoblast lines. The HSATII repeat is labeled. As in Figure 1A, to ensure all repeat types are shown on this log-scaled plot, zero values are replaced with an arbitrarily low value and points shown with an “x”. (B) HSATII is massively activated by DUX4. For our four RNA-seq samples, and sixteen “Body Map” tissues sequenced by Illumina, we show the proportion of reads that match HSATII pericentromeric satellite repeats on a log10-scale. DUX4-expressing cell lines are shown in red, and have a much higher level of HSATII expression than any other sample surveyed.
