Abstract
There is a growing appreciation that metabolic signals are integrated and coupled to cell cycle progression. However, the molecular wiring that connects nutrient availability, biosynthetic intermediates and energetic balance to the core cell cycle machinery remains incompletely understood. In this review, we explore the recent progress in this area with particular emphasis on how nutrient and energetic status is sensed within the cell to ultimately regulate cell growth and division. The role these pathways play in normal cell function including stem cell biology is also discussed. Furthermore, we describe the growing appreciation that dysregulation of these pathways might contribute to a variety of pathological conditions including metabolic diseases and tumor formation.
The creation of two cells from one requires an enormous generation of new proteins, lipids and nucleic acids. As such, the decision of a cell to enter the cell cycle represents an energetic obligation, a sort of metabolic IOU. While the general mechanisms of cell cycle checkpoints has deservedly received considerable attention, much of the emphasis has been placed on molecules such as cyclin-dependent kinases (CDKs) and their well characterized cellular partners, the cyclins. Nonetheless, how this core machinery is integrated with the cell cycle dependent biosynthetic demands of the cell has, until recently, been largely neglected. Early gene expression studies suggested that the expression of metabolic enzymes appeared to be synchronized with certain discrete phases of cell cycle progression. For instance, using S. cerevisiae as a model, it was shown that many nuclear-encoded mitochondrial enzymes required for glycolysis and oxidative phosphorylation were induced in early G1[1]. A similar pattern was observed in this study for genes involved in fatty acid biosynthesis. Related studies, again using yeast as a model, demonstrated that there was also clear cell cycle regulation of the expression of genes involved in nutrient uptake and amino acid synthesis [2]. Thus, these early transcriptome studies established the notion that the biosynthetic machinery was, as might be expected, intimately coupled and coordinated with the cell cycle.
These initial observations were further enhanced by analyzing the growth of budding yeast under culture conditions where the pH, temperature and nutrient levels could be strictly regulated. Under such conditions, yeast cultures were observed to undergo spontaneous and periodic oscillations in oxygen consumption [3]. These yeast metabolic cycles (YMC) occurred over a time scale of 4–5 hours and corresponded to periodic and coordinated gene expression of roughly half of all yeast transcripts. Of the 100 or so genes that demonstrated the greatest periodicity, nearly two thirds were involved in mitochondrial function. Further analysis of this system revealed that each phase of the observed metabolic cycle could be equated with an equivalent classical phase of the cell cycle [3]. Subsequent analysis revealed that in addition to oxygen consumption exhibiting periodicity, oscillating cycles of biosynthetic components including amino acids, nucleotides, as well as, energetic intermediates such as NADP(H) and acetyl-CoA could also be observed [4].
These studies in simple organisms such as yeast have been supplemented with additional observations in higher organisms that appear to further link metabolism with the cell cycle. Work over the last two decades demonstrated that the E2F transcription factor family is critical for the G1/S transition by regulating the expression of a host of factors including thymidylate synthase, ribonucleotide reductase and dihydrofolate reductase [5]. More recent observations have further demonstrated that mice deficient in the E2F1 transcription factor exhibit altered energy metabolism and that the cdk4-Rb-E2F transcriptional network can repress mitochondrial oxidative metabolism [6*]. In a reciprocal fashion, increased levels of mitochondrial reactive oxygen species were shown to activate E2F1 [7*]. Additional studies in mice have also linked the cdk4-Rb-E2F pathway to a host of metabolic parameters including pancreatic β-cell size and function, skeletal muscle metabolism and white adipose cell function [8]. Similarly, mice engineered to be deficient in two CDK inhibitors (p21 and p27) developed marked obesity. For instance, compared to wild type mice, p21−/−p27−/− deficient mice were noted to have a nearly fourfold increase in the percentage of body fat [9]. These observations may extend to humans, since in genome wide association studies, certain loci of CDK inhibitors have been strongly associated with susceptibility to type II diabetes [10,11].
Sensing Energy Status
The establishment of a link between metabolism and growth suggests that the cell must have a way of sensing energetic status. Indeed, the simple observation that for most cells in culture, the withdrawal of extracellular nutrients (e.g. serum) results in a corresponding withdrawal from the cell cycle demonstrates this apparent connection. Considerable progress has been made in understanding how the cell senses its underlying metabolic state and how these energetic sensors are coupled to downstream effectors of the cell cycle. The prototypical intracellular sensor is AMP-activated protein kinase (AMPK). AMPK appears to exist in all eukaryotes in the form of a heterotrimeric complex composed of a catalytic α subunit and regulatory β and γ subunits. For many years, as it name suggest, the adenine nucleotide AMP was viewed as the intracellular activator of AMPK. Recent evidence suggests however that the more abundant nucleotide ADP may be the more important regulator during most energy-depleted conditions [12,13]. Activation of AMPK initiates a host of metabolic changes that can globally viewed as an attempt to restore energy homeostasis [14]. Among these changes are phosphorylation of a myriad of downstream targets that increase the uptake of substrates (e.g. glucose and fatty acids) and the corresponding inhibition of processes that consume biosynthetic intermediates (e.g. decrease in glycogen, cholesterol and protein synthesis). Activation of AMPK also leads to the phosphorylation of the tumor suppressor gene p53 (serine-15) with the subsequent activation of p53 and the induction of a p21waf1/cip1 mediated cell cycle arrest [15]. Subsequent studies suggested that AMPK can also regulate the phosphorylation and activity of p27, providing yet another way metabolic status can be coupled to cell growth or cell fate [16]. Another important target of AMPK is the mTOR pathway. TSC2, an upstream regulator of mTOR, and Raptor, a subunit of mTORC1 are both targets of AMPK [17,18].
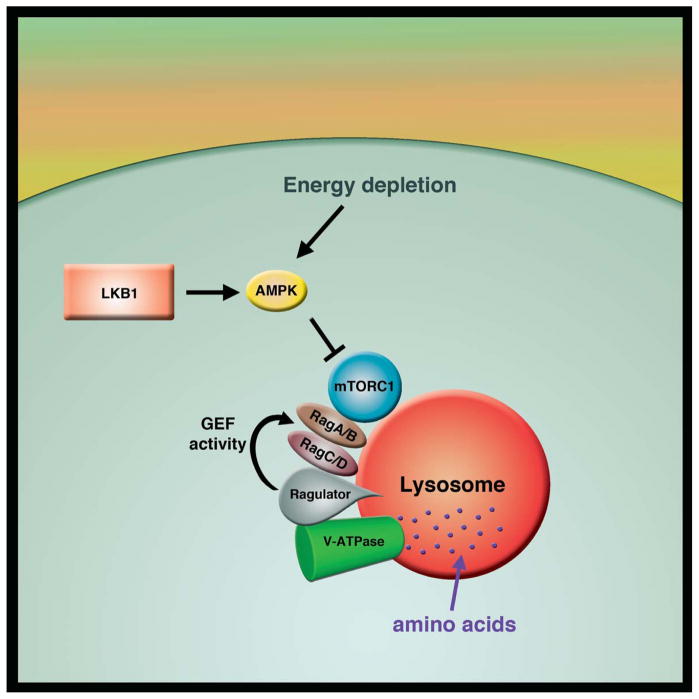
The link between AMPK and mTOR adds to the growing appreciation that mTOR is also an important sensor of overall nutrient status. The mTOR protein is a large serine/threonine kinase that exists in two distinct protein complexes termed mTORC1 and mTORC2. Extracellular growth factors such as insulin and intracellular factors such as amino acids levels appear to regulate mTOR activation. While mTOR has been known for a long time to respond to changes in amino acids such as leucine, considerable progress has been made recently on understanding the precise mechanism and location for this sensing. Evidence suggests that levels of amino acids are sensed at the surface of lysosome and that mTOR is recruited to this organelle via and interaction between the mTORC1 component Raptor and the Rag family of GTPases [19,20]. The Rag GTPases appear in turn to be anchored to the lysosomal surface by a large scaffolding complex termed the Ragulator that acts as guanine nucleotide exchange factor (GEF) for the Rag GTPases [21**]. Additional elements of this signaling pathway include the vacuolar ATPase (v-ATPase) that is essential for maintaining the low pH of the lysosome [22]. Current models suggest that a rise in intra-lysosomal amino acids can be transmitted via an inside-out mechanism through the v-ATPase, that in turn, directly interacts and alters the activity of Rag-Ragulator complex (Figure 1). While the preponderance of work has been centered on amino acid sensing, there is a growing appreciation that this pathway may also respond to other energetic stresses including low glucose [23]. Once activated, mTOR appears to have hundreds of downstream targets that promote cell growth and cell cycle progression, including a newly defined role in coordinating pyrimidine synthesis with S-phase progression [24*]. Finally, there is growing evidence that specific amino acids can also regulate cell growth in an apparent mTOR-independent fashion [25,26].
Figure 1.
Activation of mTORC1 occurs at the lysosomal surface. Amino acid levels are sensed through an inside-out mechanism using the lysosomal V-ATPase. The sensing mechanism also includes the Ragulator complex, which acts as a guanine nucleotide exchange factor (GEF) for the Rag family of GTP binding proteins. The activity of mTOR is negatively regulated by AMPK that senses AMP and ADP levels during energy depleted conditions, or is activated by a variety of other energy sensors including LKB1.
Mitochondria and the cell cycle
While the withdrawal of serum from cultured cells is well known to inhibit DNA synthesis, it is perhaps less well appreciated that reductions in extracellular amino acids, glucose or even phosphate ions can cause a G0/G1 growth arrest [27]. One mediator of this nutrient or metabolic checkpoint appears to be the previously described AMPK-dependent activation of p53-mediated growth arrest, initially described in cultured cells deprived of glucose [15]. Subsequent studies suggested this pathway could be activated by other inducers of energetic stress such as disruption of mitochondrial electron transport, and this regulation appears to be conserved in lower organisms [28]. Interestingly, in Drosophila, mitochondrial dysfunction causes cell cycle arrest by at least two distinct pathways. One involves an AMPK-dependent, p53-mediated induction of Cyclin E degradation [28,29]. The other retrograde signaling pathway leading to G1 arrest involves the mitochondrial reactive oxygen species (ROS) dependent induction of the Drosophila p27 homolog Dacapo [30]. Removal of nutrients also stimulates the induction of autophagy. How this process is coordinated with cell cycle withdrawal is incompletely understood. Recent evidence suggests however that one way these processes are coupled is through the ability of the essential autophagy gene Atg7 to directly bind p53 and modulate the induction of p21waf1/cip1 under nutrient starved conditions [31*].
In the examples mentioned above, cell cycle arrest appears to be mediated by either a decline in bioenergetics (e.g. a rise in AMP or nutrient withdrawal) or by evidence of mitochondrial stress (e.g. increased ROS levels). In yeast, however, there is evidence that a reduction of mitochondrial DNA may be sufficient to induce G1 arrest [32*]. For instance, yeast lacking mitochondrial DNA underwent cell cycle arrest, while yeast engineered to express non-coding mitochondrial DNA did not display this cell cycle defect. These observations suggest that this cell cycle arrest was caused by the absence of mitochondrial DNA and not the absence of mitochondrial-encoded gene products. Enforcement of G1 arrest in the absence of mitochondrial DNA was mediated by Rad53, the yeast ortholog of the mammalian Chk2 kinase. These observations suggest that proteins such as ATM and Chk2 that signal cell cycle arrest following DNA damage, might also play a role in regulating mitochondrial-dependent DNA maintenance and checkpoints. Consistent with such a hypothesis, there is evidence that in mice and humans, ATM is required to maintain mitochondrial DNA copy number [33]. Furthermore, deletion of Chk2 in mice appears to limit some of the phenotypic alterations induced by either mitochondrial dysfunction or nutrient stress [31*,34].
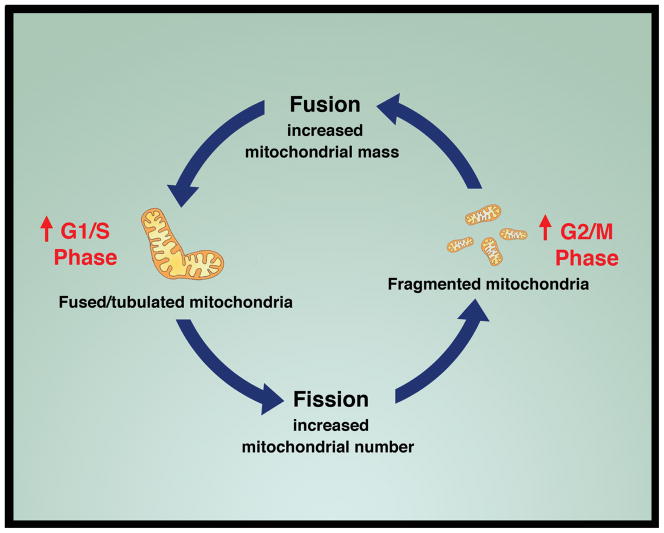
In a potentially related manner, there is a growing appreciation for the role and influence of mitochondrial dynamics in cell cycle progression. Mitochondria can exist in a fragmented state, a fused, tubulated state, or as is more common for cells growing in culture, some mixture of both mitochondrial morphologies [35]. Early observations suggested that in yeast, at the beginning of S-phase, mitochondria could fuse into a large, connected network [36]. In mammalian cells, evidence suggests that as cells progress through G1, mitochondrial membrane potential and respiration markedly increases [37]. Furthermore, careful imaging of cells demonstrated that at the G1/S boundary, mitochondria appear to undergo a marked alteration in morphology, forming a giant, hyperfused and hyperpolarized network [38]. These mitochondrial changes appear to increase bioenergetic capacity and to regulate Cyclin E accumulation. Subsequent studies have demonstrated that elements of the mitochondrial fusion machinery appear to be degraded in a cell cycle dependent fashion [39]. Taken together, these observations suggest a growing connection between the cell cycle and the morphology of mitochondria (Figure 2). This connection may extend to cell growth in general. For instance, the Hippo pathway in Drosophila is an important regulator of cell size and growth. In flies, this pathway consists of the proteins Hippo/Warts/Yorkie, with nuclear Yorkie acting as the transcriptional effector of the cellular overgrowth phenotype. Interestingly, activation of this pathway in Drosophila has been recently shown to increase mitochondrial number and augment mitochondrial fusion [40**]. This phenomenon appears to be at least partially conserved, as this study demonstrated that expression of YAP2, a mammalian ortholog of Yorkie, also appears to increase mitochondrial fusion in certain mammalian cell lines. Interestingly, other reports suggest that activation of the Hippo pathway may be a downstream effector of mitochondrial dysfunction [41**]. Given the growing link between alterations in the Hippo pathway and tumorgenesis [42], it is tempting to speculate that these recent observations connecting this pathway as both an upstream and downstream effector of mitochondrial function may at least partially explain some of the unique aspects of cancer metabolism.
Figure 2.
Mitochondrial dynamics and the cell cycle. Mitochondria can undergo profound alterations in their morphology. Fusion results in elongated, tubulated mitochondria, while fission results in mitochondria that are fragmented. Recent evidence suggests that mitochondrial morphology is coordinated with cell cycle phases with fused mitochondria occurring at the G1/S boundary and fragmented mitochondria occurring more frequently during G2/M.
Metabolism and stem cell biology
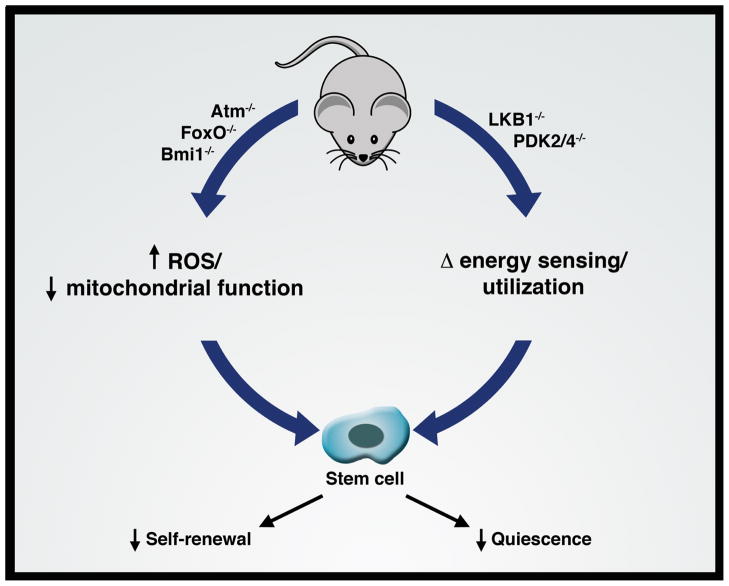
The metabolic regulation of cell cycle progression may have particular relevance in the biology of certain highly specialized cell types. The last few years has seen a particular interest in uncovering the role of metabolism in stem cell biology. Mice deficient in the ATM kinase, the Polycomb gene Bmi1, or the FoxO family of transcription factors, demonstrate an increase in ROS levels or a decline in mitochondrial function that has been linked to a corresponding alteration in the function of hematopoietic stem cells (HSCs) and/or neural stem cells [34,43–45]. Similarly, evidence suggests that HSCs conditionally deleted for LKB1 demonstrate impaired stem cell quiescence [46–48]. LKB1, an upstream regulator of AMPK, is a serine/threonine kinase that regulates cell growth in response to nutrient availability. Under normal conditions, approximately 90% of HSCs within the bone marrow are in a G0, non-cycling condition. This fraction was observed to be much higher in LKB1−/− HSCs, leading to rapid depletion of the stem cell compartment. While these effects appeared independent of AMPK and mTOR, LKB1−/− HSCs had clear alterations in mitochondrial function and bioenergetics. This suggests a poorly understood metabolic requirement to maintain stem cell quiescence (Figure 3). Interestingly, metabolic status also appears to regulate entry and exit from quiescence in yeast [49]. Recently, a metabolomic analysis of mouse HSCs revealed that bone marrow cells with long term repopulating ability exhibited increased levels of pyruvate and evidence of reduced pyruvate dehydrogenase (PDH) activity [50*]. PDH activity is normally regulated, in part, by the family of PDH kinases (PDKs). The phosphorylation of PDH by PDK family members results in a reduction of PDH activity thereby shunting carbohydrate metabolism away from the mitochondria. As such, this recent observation of decreased PDH activity is consistent with previous observations that HSCs have reduced mitochondrial metabolism and a heavy reliance on cytosolic glycolysis [51]. Furthermore, in this recent study, genetic manipulations of the PDKs impaired quiescence in HSCs, again suggesting a tight correlation between metabolism, in this case glycolytic flux, and the ability of HSCs to maintain their normally quiescent phenotype [50*]. Finally, the relationship between metabolism and cell cycle progression is not confined to stem cell biology. For instance, there is a growing appreciation that metabolic checkpoints are important regulators of immune cells [52]. Furthermore, evidence suggests that distinct metabolic programs such as cytosolic glycolysis or mitochondrial fatty acid metabolism may regulate T cell fate [53–55].
Figure 3.
Stem cell biology and metabolism. Various recent mouse models have linked the intracellular metabolism of stem cells with certain specific alterations. For instance, mice deficient in a variety of kinases (ATM), transcription factors (FoxO family), or chromatin modifiers (Bmi1) exhibit alterations in mitochondrial function or redox homeostasis. Similarly, disruption of genes involved in energy sensing (LKB1) or regulation of metabolic enzymes (PDK2 and PDK4) alter stem cell metabolism. These models in turn appear to exhibit profound defects in stem cell function including alterations in stem cell self-renewal capacity or maintenance of quiescence.
Concluding Remarks and Future Perspectives
In conclusion, while incompletely understood, cell cycle progression is tightly coupled to intracellular metabolism. Emerging evidence suggests that intracellular kinases such as AMPK and mTOR can sense energy intermediates such as AMP, ADP and amino acids and subsequently direct how biosynthetic intermediates are used and whether the cell should arrest, grow or divide. Mitochondrial function and morphology is also apparently coupled to cell cycle dynamics with fused mitochondria apparently essential for the G1/S transition. Perhaps even more exciting, we are beginning to see the outlines as to how metabolism may be uniquely altered and regulated within specialized cells. In this regard, studies with various stem cell populations and immune cells hint that biasing the cell towards specific metabolic pathways can in turn alter cell growth and fate. Therapies aimed at modulating metabolism therefore holds promise as a means to alter stem cell behavior and function, as well as augmenting or dampening the immune response. While the health benefits of caloric restriction are well known, a deeper understanding of intracellular bioenergetics may allow targeted interventions that directly modulate specific metabolic pathways in key regulatory cell types. Furthermore, a deeper understanding of these pathways may provide insight into a host of human diseases. For instance, it seems increasingly likely that there is an important mechanistic link between the dysregulation of cell cycle checkpoints and the alterations in metabolism often found together within tumor cells. Whether the metabolic changes are primary or secondary, and whether these changes are upstream, downstream or independent of the observed alterations in cell cycle parameters remains largely unknown. Nonetheless, the recent renewed interest in metabolism suggests that many of these questions should soon have answers.
Acknowledgments
We are grateful to Ilsa Rovira for help with the figures. This work was supported by NIH Intramural funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cho RJ, Campbell MJ, Winzeler EA, Steinmetz L, Conway A, Wodicka L, Wolfsberg TG, Gabrielian AE, Landsman D, Lockhart DJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 2.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 4.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nevins JR. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- *6.Blanchet E, Annicotte JS, Lagarrigue S, Aguilar V, Clape C, Chavey C, Fritz V, Casas F, Apparailly F, Auwerx J, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol. 2011;13:1146–1152. doi: 10.1038/ncb2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *7.Raimundo N, Song L, Shutt TE, McKay SE, Cotney J, Guan MX, Gilliland TC, Hohuan D, Santos-Sacchi J, Shadel GS. Mitochondrial stress engages E2F1 apoptotic signaling to cause deafness. Cell. 2012;148:716–726. doi: 10.1016/j.cell.2011.12.027. These two studies place the E2F1 transcription factor both upstream and downstream of mitochondria. The latter study further suggests this pathway may be important for age-dependent hearing loss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguilar V, Fajas L. Cycling through metabolism. EMBO Mol Med. 2010;2:338–348. doi: 10.1002/emmm.201000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naaz A, Holsberger DR, Iwamoto GA, Nelson A, Kiyokawa H, Cooke PS. Loss of cyclin-dependent kinase inhibitors produces adipocyte hyperplasia and obesity. FASEB J. 2004;18:1925–1927. doi: 10.1096/fj.04-2631fje. [DOI] [PubMed] [Google Scholar]
- 10.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 11.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332:1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- 13.Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472:230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardie DG, Ross FA, Hawley SA. AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Biol. 2012;19:1222–1236. doi: 10.1016/j.chembiol.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- 17.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 18.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **21.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. This manuscript provides crucial molecular details as to how amino acid levels are sensed by the mTOR kinase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. The presumed multitude of downstream targets of mTOR are largely unknown. Using newer proteomic techniques these investigators link mTOR to pyrimidine synthesis suggesting a way nutrient sensing can be coupled to S phase progression. [DOI] [PubMed] [Google Scholar]
- 25.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holley RW, Kiernan JA. Control of the initiation of DNA synthesis in 3T3 cells: low-molecular weight nutrients. Proc Natl Acad Sci U S A. 1974;71:2942–2945. doi: 10.1073/pnas.71.8.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Mandal S, Freije WA, Guptan P, Banerjee U. Metabolic control of G1-S transition: cyclin E degradation by p53-induced activation of the ubiquitin-proteasome system. J Cell Biol. 2010;188:473–479. doi: 10.1083/jcb.200912024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- *31.Lee IH, Kawai Y, Fergusson MM, Rovira, Bishop AJ, Motoyama N, Cao L, Finkel T. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–228. doi: 10.1126/science.1218395. Nutrient withdrawal induced both cell cycle arrest and the induction of autophagy. This manuscript suggests one mechansim for how these two events might be coordinated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.Crider DG, Garcia-Rodriguez LJ, Srivastava P, Peraza-Reyes L, Upadhyaya K, Boldogh IR, Pon LA. Rad53 is essential for a mitochondrial DNA inheritance checkpoint regulating G1 to S progression. J Cell Biol. 2012;198:793–798. doi: 10.1083/jcb.201205193. This manuscript demonstrates that in yeast, the presence of mitochondrial DNA is required for cell cycle progression. Interestingly, this effect does not appear to work through a bioenergetic mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eaton JS, Lin ZP, Sartorelli AC, Bonawitz ND, Shadel GS. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Invest. 2007;117:2723–2734. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 36.Miyakawa I, Aoi H, Sando N, Kuroiwa T. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J Cell Sci. 1984;66:21–38. doi: 10.1242/jcs.66.1.21. [DOI] [PubMed] [Google Scholar]
- 37.Schieke SM, McCoy JP, Jr, Finkel T. Coordination of mitochondrial bioenergetics with G1 phase cell cycle progression. Cell Cycle. 2008;7:1782–1787. doi: 10.4161/cc.7.12.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park YY, Cho H. Mitofusin 1 is degraded at G2/M phase through ubiquitylation by MARCH5. Cell Div. 2012;7:25. doi: 10.1186/1747-1028-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Nagaraj R, Gururaja-Rao S, Jones KT, Slattery M, Negre N, Braas D, Christofk H, White KP, Mann R, Banerjee U. Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev. 2012;26:2027–2037. doi: 10.1101/gad.183061.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **41.Ohsawa S, Sato Y, Enomoto M, Nakamura M, Betsumiya A, Igaki T. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature. 2012;490:547–551. doi: 10.1038/nature11452. Two interesting manuscripts that link the Hippo pathway with mitochondrial function. Again, there appears to be a role for this pathway both upstream and downstream from mitochondria. May ultimately have important implications for normal cell growth and for tumor formation. [DOI] [PubMed] [Google Scholar]
- 42.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 43.Ahlqvist KJ, Hamalainen RH, Yatsuga S, Uutela M, Terzioglu M, Gotz A, Forsstrom S, Salven P, Angers-Loustau A, Kopra OH, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 46.Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, Fletcher-Sananikone E, Colla S, Wang YA, Chin L, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J Cell Biol. 2011;192:949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *50.Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. This manuscript reveals a mechanism through which metabolic activity is linked to stem cell quiescence. Demonstrates growing interest in better understanding how intracellular metabolic choices regulate stem cell fate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, Sadek HA. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 53.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]