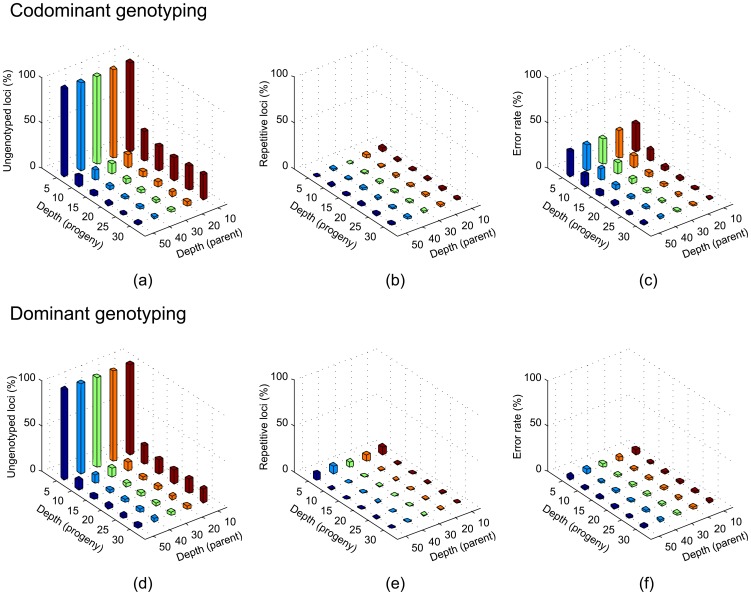
Figure 2. Evaluation of the performance of RADtyping using a pseudo F1 mapping population.
The simulated population was created by a crossing of two Arabidopsis plants with predefined SNPs in their genomes and progeny were subject to in silico sequencing together with their parents at different sequencing depths with sequencing errors enabled. De novo codominant and dominant genotyping was evaluated in three key aspects: genotype coverage (a, b), removal of repetitive sites (b, e), and genotyping accuracy (c, f).