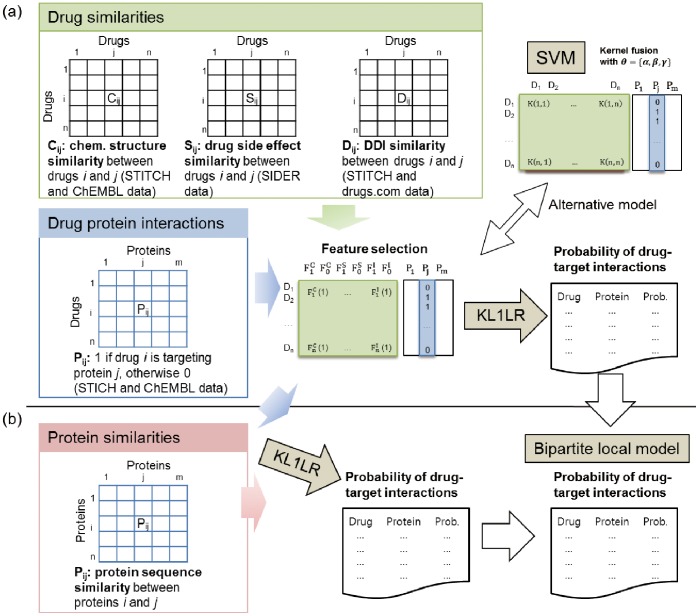
Figure 2. Overview of the entire method procedure.
(a) Drug similarities and drug-protein interactions are used to calculate the probabilities of unknown drug-target interactions. Three different drug similarities (chemical structure, drug side effect, and DDI similarity) are applied. Two learning models (KL1IR and SVM) are used to train and test interactions. (b) Protein similarities are integrated with drug similarities to predict unknown drug-target interactions. In this process, the bipartite local model is used.