Table 2. Comparison of 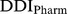 and KEGG pharmacological information.
and KEGG pharmacological information.
| Methods | Data Sources | Kernels |
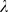 or C or C |
AUC |
| KL1LR | STITCH | DDIPharm | 0.04 | 0.7508 |
| KL1LR | STITCH | Pharmacology | 0.04 | 0.7369 |
| KL1LR | KEGG | DDIPharm | 0.04 | 0.7316 |
| KL1LR | KEGG | Pharmacology | 0.03 | 0.7232 |
| SVM | STITCH | DDIPharm | 2 | 0.7982 |
| SVM | STITCH | Pharmacology | 0.75 | 0.7685 |
| SVM | KEGG | DDIPharm | 30 | 0.6727 |
| SVM | KEGG | Pharmacology | 20 | 0.7089 |
Two sets of drug-target interactions from STITCH and KEGG were used for prediction. 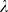 and
and  are the KL1LR and SVM parameters, respectively; Table S1 contains prediction results when different parameter values were used. Table S2 contains ROC curves of true positive rate and false positive rate, and tables of true positive, false positive, true negative, and false negative values for each threshold.
are the KL1LR and SVM parameters, respectively; Table S1 contains prediction results when different parameter values were used. Table S2 contains ROC curves of true positive rate and false positive rate, and tables of true positive, false positive, true negative, and false negative values for each threshold.
