Table 5. Comparison of drug-target interactions predictions from experiments and those from experiments and text mining in STITCH.
| Method | Source of drug-target interaction | Chemical structure | Side effect | DDIPharm | CS | CSD |
| KL1LR | Experiments only | 0.7800 | 0.7744 | 0.8311 | 0.8221 | 0.8288 |
| KL1LR | All | 0.7787 | 0.7768 | 0.8294 | 0.7774 | 0.8267 |
| SVM | Experiments only | 0.8149 | 0.8360 | 0.8174 | 0.8410 | 0.8461 |
| SVM | All | 0.7682 | 0.7820 | 0.8009 | 0.7843 | 0.8078 |
AUC values for predicting drug-target interactions are shown to compare two cases of using experimentally validated interactions and by using all interactions including experiments, text mining, and other databases. The comparison was conducted using two prediction models (KL1LR and SVM) and three drug similarities (chemical structure, side effect, and 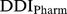 ) and combining them. CS and CSD indicate the drug similarity by combining CH and SE, and combining CH, SE, and DDI, respectively. For the choice of parameter values, see Table S6.
) and combining them. CS and CSD indicate the drug similarity by combining CH and SE, and combining CH, SE, and DDI, respectively. For the choice of parameter values, see Table S6.
