Abstract
Non-invasive imaging holds significant potential for implementation in tissue engineering. It can e.g. be used to monitor the localization and function of tissue-engineered implants, as well as their resorption and remodelling. Thus far, however, the vast majority of efforts in this area of research have focused on the use of ultrasmall super-paramagnetic iron oxide (USPIO) nanoparticle-labeled cells, colonizing the scaffolds, to indirectly image the implant material. Reasoning that directly labeling scaffold materials might be more beneficial (enabling imaging also in case of non-cellularized implants), more informative (enabling the non-invasive visualization and quantification of scaffold degradation) and more easy to translate into the clinic (since cell-free materials are less complex from a regulatory point-of-view), we here prepared three different types of USPIO nanoparticles, and incorporated them both passively and actively (via chemical conjugation; during collagen crosslinking) into collagen-based scaffold materials. We furthermore optimized the amount of USPIO incorporated into the scaffolds, correlated the amount of entrapped USPIO with MR signal intensity, showed that the labeled scaffolds are highly biocompatible, demonstrated that scaffold degradation can be visualized using MRI and provided initial proof-of-principle for the in vivo visualization of the scaffolds. Consequently, USPIO-labeled scaffold materials seem to be highly suitable for image-guided tissue engineering applications.
Keywords: Imaging, Tissue Engineering, Collagen Scaffolds, Iron oxide nanoparticles, MRI
1. Introduction
Tissue engineering refers to the use of biological, chemical and nanotechnological materials to replace or restore the function of damaged organs and tissues.[1;2] Many different tissue-engineered implants have been designed and evaluated over the years, for various different purposes, including e.g. vascular grafts, ventricular valves and bone replacements.[3-6] Only very few, however, have thus far managed to successfully progress through clinical trials.[7;8] Apart from the complexity and the multifaceted nature of tissue-engineered materials, this is due - to a significant extent - to the lack of non-invasive imaging methods to longitudinally monitor their fate and function upon implantation. Here, we have therefore set out to label collagen-based tissue engineering scaffolds with ultra-small superparamagnetic iron oxide (USPIO) nanoparticles,[9;10] in order to facilitate their implantation, non-invasively assess their in vivo localization and function, and monitor their resorption. These efforts contribute to the development of improved and patient-optimized implants, they decrease the costs and the number of animals needed in preclinical trials, and they facilitate the clinical translation of tissue-engineered scaffold materials.
Magnetic resonance imaging (MRI) is highly suited for monitoring tissue-engineered implants, because of its high resolution, its high versatility and its excellent soft-tissue contrast. Among the very few efforts undertaken thus far in which MRI was used for implant imaging exclusively are studies in which USPIO-labeled cells are used to colonize scaffolds.[11;12] This approach enables the indirect imaging of scaffolds during and after implantation, and it provides information on cell survival, retention and/or replacement. However, to directly visualize implants, to also enable imaging in case of non-cellularized implants, and to obtain non-invasive imaging information on implant resorption and remodelling, it is necessary to directly label the scaffold materials. We here describe several different USPIO-based nanoparticles, scaffold-labeling techniques and MR imaging methods for the direct visualization of tissue-engineered implants, and provide initial proof-of-principle for proper imaging properties and biocompatibility.
2. Results and Discussion
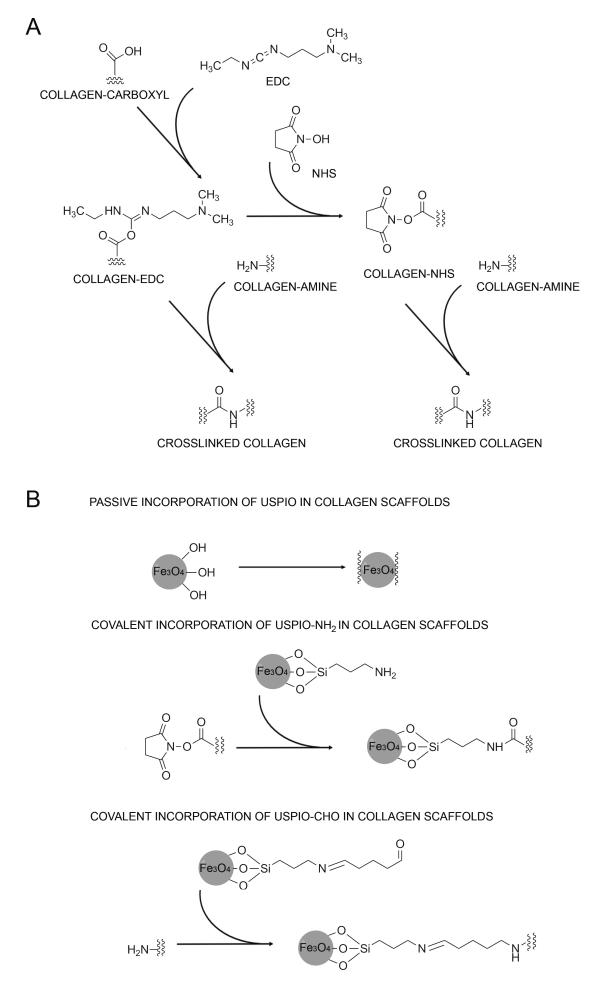
Three-dimensional collagen scaffolds, which have been extensively used for tissue engineering purposes,[13-19] were used as prototypic scaffold materials. The pores and fibers in collagen scaffolds enable cell migration into the matrix, support cellular proliferation and maintain cell viability, via the effective transport of oxygen, nutrients and metabolic waste products in and out of these materials. The typical directional pore structure of the scaffolds is obtained upon an unidirectional freezing process.[20] After freeze-drying, they are chemically crosslinked, to increase mechanical stability and resistance to enzymatic degradation.[21] As depicted schematically in Figure 1A, collagen scaffolds are crosslinked using N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide hydrochloride (EDC-HCl), which covalently connect carboxyl to amine groups, thereby forming stable amide bonds.
Figure 1. Synthesis of USPIO-labeled collagen scaffolds.
A) Chemical crosslinking of collagen scaffolds. B) Incorporation of USPIO into collagen scaffolds. USPIO possessing different surface functionalities were incorporated both passively (physical entrapment) and actively (covalent attachment) into the scaffolds.
USPIO possessing different surface functionalities were prepared, to enable both passive and active incorporation into the implant materials (Figure 1B). Non-functionalized USPIO, possessing terminal hydroxyl groups, were passively entrapped during scaffold crosslinking. APTMS (i.e. (3-aminopropyl)trimethoxysilane–tetramethoxysilane) -coated USPIO, possessing terminal amine groups, were conjugated to NHS-functionalized collagen during crosslinking. Analogously, glutaraldehyde-coated USPIO, possessing terminal aldehyde groups, were covalently attached to amine groups in the scaffolds during crosslinking.
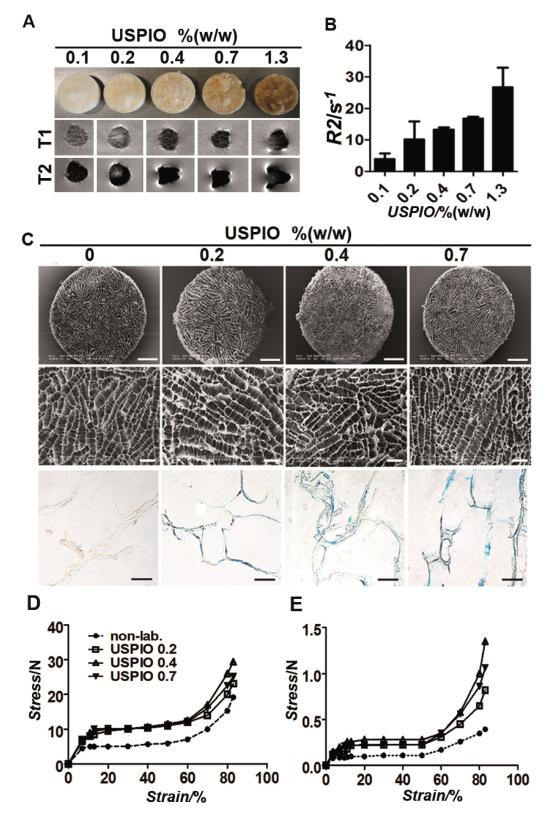
The optimal USPIO concentration for efficient MR visualization was determined by passively incorporating different amounts of USPIO, ranging from 0.1 to 1.3% (w/w), into the scaffolds. The labeled materials were then visualized using T1- and T2-weighted MRI (Figure 2A), and the corresponding R2 values were calculated on the basis of relaxometry measurements (Figure 2B). A significant increase in R2 (vs. unlabeled scaffolds) could be detected at concentrations of 0.2% (w/w) and beyond. In addition, at concentrations higher than 0.7% (w/w), magnetic susceptibilities started to become very apparent (i.e. blooming effects; resulting in a deformation of the MR image; see lower panels in Figure 2A), making an accurate size determination of the scaffold and a proper interpretation of the area surrounding the scaffold increasingly complicated. Consequently, USPIO amounts of 0.2-0.7% (w/w) resulted in the most optimal imaging properties, providing sufficient contrast enhancement for visualizing and sizing the scaffolds, while keeping the impact of magnetic susceptibilities reasonably small.
Figure 2. MR analysis and structural characterization of collagen scaffolds labeled with increasing amounts of USPIO.
A) Visual depiction and T1- and T2-weighted MR images of scaffolds containing passively entrapped USPIO at iron concentrations ranging from 0.1-1.3 % (w/w). B) Quantitative R2-relaxometry analysis. C) Low and high resolution SEM images and Prussian Blue stainings of labeled scaffolds, showing that USPIO-incorporation does not affect the pore structure of the materials (see Table S1 for quantification). Scale bars indicate 1 mm, 200 μm and 100 μm in the top, middle and bottom panel, respectively. D-E) Compression testing of non-labeled and labeled collagen scaffolds in dry (D) and wet (E) state, indicating an increase in mechanical stiffness upon USPIO incorporation.
Upon having identified the most optimal USPIO concentrations for implant imaging, the scaffolds were labeled both passively and actively (Figure 1B), and their structural properties, labeling stability, MR visibility and biocompatibility were evaluated. Scanning electron microscopy (SEM) demonstrated that the pore structure of the scaffolds was not affected by the incorporation of USPIO (Figure 2C). The pore sizes of the labeled scaffolds were comparable to those of non-labeled scaffolds, and were in the range of 80 to 100 μm (Table S1). Also the collagen helix denaturation temperature, determined using dynamic scanning calorimetry (DSC), was unaffected by USPIO labeling (Table S1). The presence of incorporated iron in the implant materials was confirmed using Prussian Blue Staining (Figure 2C) and ICP-MS analysis (Figure S1). USPIO labeling was found to be highly stable, and to be independent of the mode of incorporation: neither for passively entrapped USPIO, nor for chemically incorporated USPIO, a significant loss of contrast agent was observed up until 4 weeks of extensive washing (Figure S1).
The mechanical properties of the labeled scaffolds were assessed via compression testing. The compression curves obtained for all samples displayed the typical shape of compression curves for foam-like materials (Figure 2D-E). Three different types of material response were observed: linear elasticity, long flat plateau and finally densification of the material, causing a sharp stress raise. USPIO-labeled scaffolds in dry (Figure 2D) and wet (Figure 2E) state showed an increase in compressive modulus, indicating an enhancement of mechanical stiffness upon nanoparticle incorporation. The stress at the maximal scaffold compression of 83% in dry state was 19.2±0.7 N for non-labeled scaffolds, 23.1±1.1 N for scaffolds with 0.2% (w/w) USPIO, 29.4±2.0 N for scaffolds with 0.4% (w/w) USPIO and 25.1±1.1 N for scaffolds with 0.7% (w/w) USPIO. Stress values for the same materials in wet state were 0.39±0.02 N, 0.82±0.04 N, 1.35±0.11 N and 1.06±0.10 N, respectively. These slight but significant increases (r2 correlation coefficients: 0.9072 (dry), 0.9607 (wet), p-values: < 0.001) in mechanical properties might be beneficial for implants which are exposed to moderate to high mechanical stress, but they also have to be carefully taken into account when tailoring the resorption and degradation rates of the scaffolds.
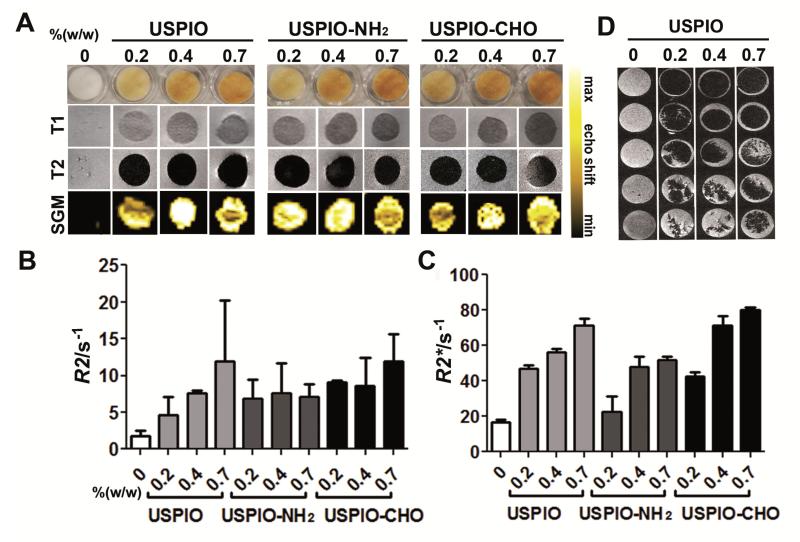
Subsequently, crosslinked collagen scaffolds passively and actively labeled with 0.2-0.7% (w/w) USPIO, USPIO-NH2 and USPIO-CHO were visualized and analyzed using MRI. As shown in Figure 3A, all three labeled scaffold materials could be clearly identified in T1- and T2-weighted MR imaging, whereas non-labeled scaffolds could not be detected. Contrast-to-noise ratios (CNR) in T1-weighted images increased from 0.85±0.70 for non-labeled scaffolds to 9.10±1.05 for 0.2% (w/w) USPIO, 9.60± 0.81 for 0.4% (w/w) USPIO and 17.63±0.68 for 0.7% (w/w) USPIO; and in T2-weighted images from 0.63±0.32 for non-labeled scaffolds to 13.69±1.67 for 0.2% (w/w) USPIO, 15.21±0.81 for 0.4% (w/w) USPIO and 19.44±0.14 for 0.7% (w/w) USPIO. Quantitative R2 and R2* relaxometry analyses correlated relatively well with the degree of USPIO incorporation (Figure 3B-C). Strikingly, in particular for passively incorporated USPIO, a linear relationship and high correlation between the amount of incorporated iron and R2 relaxation rate was observed (r2 correlation coefficients and p-values: 0.9479 and <0.0001 for USPIO, vs. 0.6526 and 0.0084 for USPIO-NH2, and vs. 0.8180 and 0.0008 for USPIO-CHO, respectively).
Figure 3. MR imaging of collagen scaffolds labeled both passively and actively with USPIO.
A) Visual depiction, T1- and T2-eighted MRI, and Susceptibility Gradient Mapping (SGM) of labeled scaffolds. B-C) Quantitative R2- (B) and R2*-relaxometry (C) analysis. All labeled scaffolds were clearly detectable in MRI and showed significant increases in R2 and R2* relaxation rates with increasing amounts of incorporated USPIO. D) MR monitoring of scaffold degradation over time upon exposure to collagenase.
As the negative contrast of USPIO can be confused with other sources of negative contrast, such as air, we also performed bright iron imaging, using a Susceptibility Gradient Mapping (SGM) tool. As exemplified by the lower panel images in Figure 3A, all three labeled scaffold materials now presented with hyperintense signals, which again correlated relatively well with the amount of USPIO incorporated.
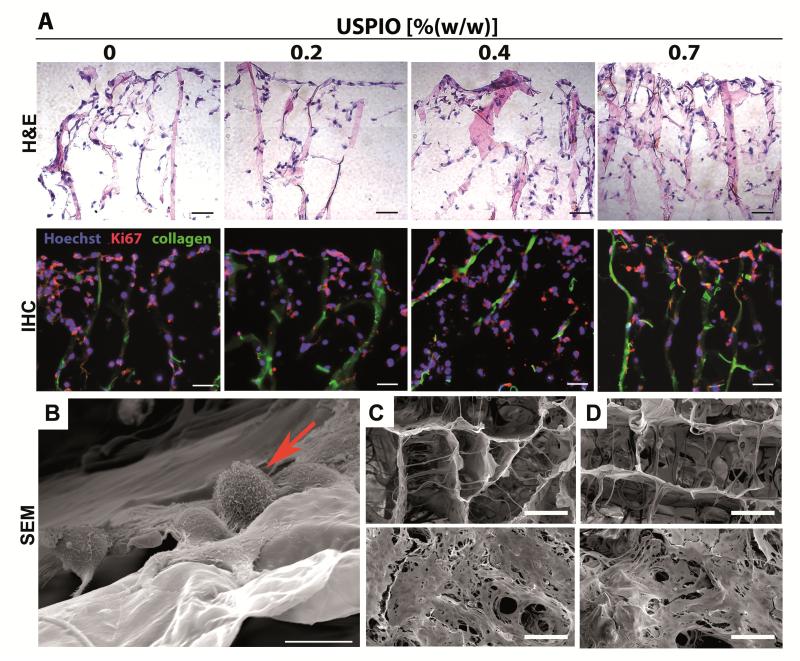
Initial proof-of-principle for using MRI to monitor the degradation of the labeled scaffold materials was obtained upon exposing them to collagenase for 1-5 days, and upon then embedding and scanning them in gelatin phantoms. As shown in Figure 3D, as expected, clear indications for a progressive degradation of the materials over time could be obtained. Next, the biocompatibility of the labeled scaffold materials was evaluated. This was done by colonizing them with three different types of physiologically relevant cells, i.e. fibroblasts, smooth muscle cells and endothelial cells. As exemplified by Figure 4A, neither the number and distribution, nor the proliferation and penetration of the cells were affected by the presence of USPIO. This notion was quantitatively confirmed by immunohistochemical and immunofluorescence analyses, in which cell density and cellular colonization were evaluated at ten different penetrations depths in three different scaffolds per group, showing that both the total number of cells and the number of Ki67-positive cells (representing proliferating cells) were comparable for each segment in labeled vs. non-labeled scaffolds (Figure S2). SEM imaging of the scaffolds, and analysis of proliferating (Ki67) and apoptotic (TUNEL) cells after 1 day and 8 days of fibroblast colonization, demonstrated the development of a dense tissue structure, both on non-labeled and labeled scaffold materials, with no apparent qualitative and quantitative differences in cell shape and viability (Figure 4B-D, Figure S3). These findings convincingly demonstrate that incorporating USPIO in collagen scaffolds does not negatively affect their ability to be used for tissue engineering purposes.
Figure 4. In vitro colonization of USPIO-labeled collagen scaffolds.
A) Light and fluorescence microscopy analysis of non-labeled and USPIO-labeled collagen scaffolds upon 8 days of colonization with NIH3T3 fibroblasts. Cells were stained with hematoxylin and eosin (H&E), with Hoechst (blue; nuclei) and with antibodies against the proliferation marker Ki67 (red). Collagen autofluorescence is depicted in green. Scale bars indicate 100 μm. No qualitative and quantitative differences in colonization were observed for labeled vs. non-labeled scaffolds (for none of the three cell types evaluated; see Figure S2). B) High-resolution SEM image of fibroblasts (arrow) colonizing a collagen scaffold. Scale bar indicates 10 μm. C-D) SEM images of the pore structure of non-labeled (C) and labeled (D) scaffolds upon 1 (top) and 8 days (bottom) of colonization with NIH3T3 fibroblasts (see Figure S3 for quantification). Scale bars indicate 100 μm.
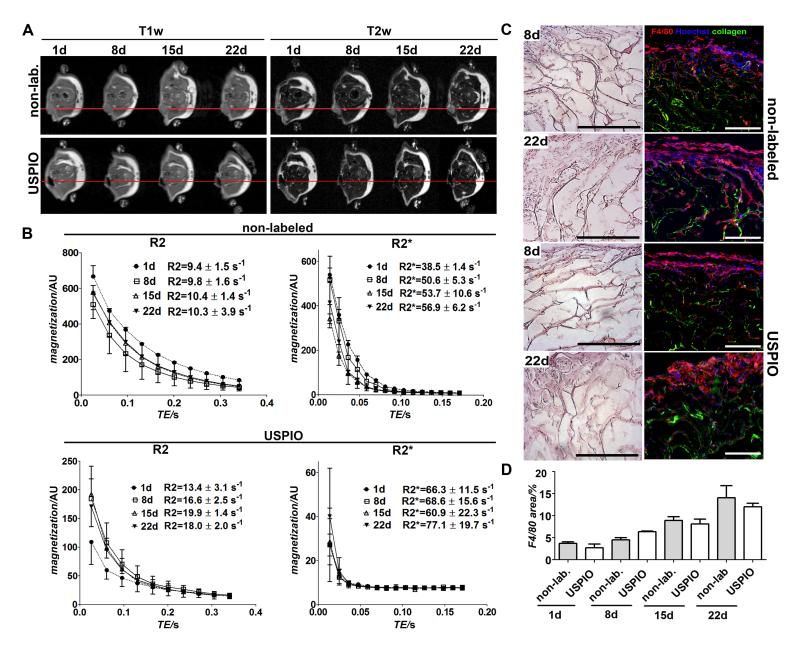
In the final set of experiments, initial proof-of-principle for monitoring the in vivo localization of USPIO-labeled collagen scaffolds was provided. As shown in Figure 5A, upon subcutaneous implantation into immunocompetent B6 mice and non-invasive imaging follow-up for up to 22 days, USPIO-labeled scaffolds could be accurately localized and sensitively visualized using T1 and T2-weighted MRI. R2 and R2* values of the regions-of-interest (ROI) covering the scaffolds were calculated on the basis of relaxometry measurements, and showed significantly higher values for labeled than for non-labeled scaffolds (Figure 5B), e.g. at day 22, R2 for labeled scaffolds was 18.0±2.0 s−1 (vs. 10.3±3.9 s−1 for non-labeled scaffolds) and R2* 77.1±19.7 s−1 (vs. 56.9±6.2 s−1 for non-labeled scaffolds).
Figure 5. In vivo evaluation of non-labeled and USPIO-labeled and collagen scaffolds.
A) T1-and T2-weighted MRI of subcutaneously implanted scaffolds over a time period of 22 days. The images clearly show that the labeled scaffolds could be sensitively detected and accurately localized. B) Quantitative analysis by R2 and R2* relaxometry. C) Light and fluorescence microscopy analysis of non-labeled and USPIO-labeled collagen scaffolds ex vivo. Cells were stained with hematoxylin and eosin (H&E), with Hoechst (blue; nuclei) and with antibodies against the macrophage marker F4/80 (red). Collagen autofluorescence is depicted in green. Scale bars indicate 200 μm. D) Quantitative evaluation of the macrophage staining by area fraction analysis. No qualitative and quantitative differences in scaffold acceptance and macrophage infiltration were observed for labeled vs. non-labeled scaffolds.
Relaxation rates at day 1 post implantation for both non-labeled and labeled scaffolds were lower compared to later time points, which can be attributed to the fact that the scaffolds were soaked in NaCl solution prior to implantation, which increases the relaxation times before it could be replaced with body liquor. At days 8, 15 and 22 post implantation, the relaxation rates showed no significant differences. The relatively constant MRI signal indicated no degradation of the scaffold material in the 22 time course, which was also confirmed by size measurements and histological analysis (Figure 5C). The collagen scaffold structure was still intact, but was found to be progressively overgrown with endogenous cells in the border areas. Macrophages were identified by F4/80 staining indicating no quantitative differences in macrophage infiltration into non-labeled or labeled scaffold tissue (Figure 5D).
Taken together, we here show that collagen-based scaffold materials can be easily and efficiently loaded with USPIO nanoparticles, thereby enabling the direct imaging of scaffold materials. Direct scaffold labeling has several distinct advantages over labeling the cells grafted onto them, including the ability to image cell-free implants and scaffold resorption, and it might also be beneficial for translation into the clinic, since cell-free materials are less complex from a regulatory point of view. In currently ongoing follow-up analyses, attempts will be made to confirm and extend these findings, not only using collagen-based implants, but also several other types of synthetic scaffolds, such as PVDF- and PLA-based fibers, meshes and grafts, in order to demonstrate in long-term in vivo studies in large animals (sheep), that the localization, resorption and function of USPIO-labeled tissue engineering materials can be non-invasively visualized using MRI.
3. Conclusion
In conclusion, we here show that collagen-based scaffold materials can be easily, efficiently and stably labeled with USPIO, and that contrast agent incorporation does not alter the ability of these materials to be used for tissue engineering applications. Neither the structural properties of the scaffolds, nor their colonization with different types of cells were negatively affected by USPIO labeling. Both passive (physical) and active (chemical) strategies for USPIO incorporation were evaluated. While both strategies resulted in highly stable incorporation, the former was found to provide the most accurate information on the amount of USPIO embedded within the scaffolds. Consequently, passively incorporating USPIO into collagen-based scaffold materials seems to be a simple and straightforward means for non-invasively visualizing the localization, resorption and function of tissue-engineered implants.
4. Experimental Section
Materials
Ferric chloride (FeCl3), ferrous chloride tetrahydrate (FeCl2.4H2O), IDRANAL III standard solution (EDTA-Na2; reagent for metal titration; 0.1 M), toluene, 3-aminopropyltrimethoxysilane (APTMS), glutaraldehyde and Nuclear Fast Red Solution were purchased from Sigma Aldrich (Steinheim, Germany). Ammonia, hydrochloric acid, ammonium peroxodisulfate, sodium hydroxide and methanol were obtained from Carl Roth (Karlsruhe, Germany). PBS buffer, gelatine and potassium ferrocyanid (K4Fe(CN)6) were acquired from AppliChem (Darmstadt, Germany). Hoechst was procured from Merck KGaA (Darmstadt, Germany) and Ki67 antibody from Novus Biological (#NB500-170). All chemicals were of appropriate analytical grade and were used without further purification.
Synthesis of USPIO
Ultrasmall super-paramagnetic iron oxide (USPIO) nanoparticles were synthesized by co-precipitation of ferrous (Fe2+) and ferric (Fe3+) salts under alkaline aqueous conditions. A modified one-pot synthetic protocol was followed. A stoichiometric ratio of 2Fe3+:Fe2+ (i.e. 12.5 mmol (2.03 g) of FeCl3 and 6.25 mmol (1.24 g) of FeCl2.4H2O), was dissolved in 190 mL de-ionized water. Afterwards, 10 mL of 25% NH3 were added under vigorous stirring and heated to 80°C. After 20 min, the particles were washed three times with 30 mL of water and two times with 30 mL of 0.1 M HCl solution by magnetic separation using a rare-earth magnet. Then, the magnetic fluids were dispersed in 25 mL of methanol. The USPIO were sonicated for 20 min prior to centrifugation at 20000 g for 15 min. The total iron concentration of the prepared USPIO was 137 mM, as determined by titrimetry.
Synthesis of USPIO-NH2
USPIO nanoparticles were dispersed in a 100 mL mixture of toluene and methanol (70:30). The suspension was then transferred into a three-necked flask equipped with a mechanical stirrer, 0.1 mL of APTMS (i.e. (3-aminopropyl)trimethoxysilane–tetramethoxysilane) was added, and the suspension was stirred under reflux at 110 °C for 10 h with nitrogen as a covering gas. The coated particles were retrieved with a magnet and dispersed in methanol by ultrasonication. Afterwards, the USPIO-NH2 particles were first washed with methanol, then with water, and finally dispersed in water. The final concentration was 135 mM.
Synthesis of USPIO-CHO
APTMS-coated USPIO were dispersed in 2 mL of 5% (v/v) glutaraldehyde in 0.1 M PBS (pH 7.4) and vacillated for 4 h at room temperature. The particles were recovered and washed three times with 0.1 M PBS, before being redispersed in 0.2 mL of PBS. The final concentration of the USPIO-CHO dispersion was 139 mM.
Synthesis and characterization of USPIO-labeled collagen scaffolds
Collagen was isolated from porcine tissues harvested under veterinary supervision from animals declared “fit for human consumption”. Tissues were purified by a series of proprietary mechanical and chemical purification steps. The purified insoluble collagen fiber network, containing 10-15% (w/w) elastin fibers, was reduced to prepare a 1.5 wt% aqueous dispersion containing insoluble fiber fragments. The dispersion was mixed for 30 min, with an appropriate amount of the different USPIO nanoparticles (see Table S2), until a homogenous brown color was obtained. The final dispersion was deaerated under vacuum.
Preparation of collagen sponges with parallel-oriented pores was based on a patented unidirectional freezing process, followed by freeze-drying. The dispersion was cooled and frozen using a defined temperature gradient and a constant cooling rate. The temperature gradient was induced using 2 heat sinks placed above and below the sample. By establishing homogeneous temperatures at the respective heat sink, a 1D heat flow was created in the direction of the temperature gradient, contrary to the direction of ice crystal growth. By cooling down the heat sinks at the beginning of the freezing process, a so-called constitutional supercooling can be induced ahead of the planar ice front. Because of this, the system becomes unstable, and by applying appropriate freezing parameters, the planar ice front can be broken down into a dendritic ice crystal morphology without side branches. After freezing, sublimation of this ice crystal structure resulted in collagen sponges with a directional pore structure. Scaffolds were crosslinked with 0,3 mM EDC (for synthesis, Merck) for 18 hours at room temperature. Sterility of the scaffolds was achieved by 25kGy gamma irradiation.
Characterization of the scaffolds
The pore size of the scaffolds was determined using a light microscope (Nikon Eclipse TE 2000-S, Nikon GmbH, Düsseldorf, Germany) by evaluating three separate slices obtained from the scaffolds (slice thickness <0.5 mm). All pores in one screen (magnification 40x) were measured using the LUCIA Measurement 4.82 image analysis software (Nikon, Leiderdorp, the Netherlands), using a calibration grid with 100 μm quadrants. For each quadrant, the most prominent pore was measured, yielding at least 100 individual measurements per screen. The three data sets obtained for the three seperate slices were combined to calculate the average pore size and the cumulative pore size distribution. The denaturation temperature of the scaffolds was determined by differential scanning calorimetry (DSC), using a TA Instrument Q100. In an empty hermetic pan, ~1 mg of scaffold was weighed, followed by the addition of 11 mg PBS. The sample was allowed to rehydrate overnight at room temperature before it was scanned at 5°C/min in the range of 15°C to 95°C. The peak temperature was taken as the denaturation temperature.
For scanning electron microscopy (SEM), the scaffolds were fixed on SEM stubs, sputter-coated with gold, and analyzed using a field emission SEM microscope (ESEM XL 30 FEG, FEI, Philips, Eindhoven, The Netherlands) in a high-vacuum environment.
Stability test
Samples of the non-labeled and USPIO-labeled scaffolds (13 mm diameter) were incubated in a phosphate buffered saline solution (pH 7,4) with 1% (v/v) streptomycin (Biochrom AG) for 4 weeks at 37°C. Samples were taken every week and were washed three times with distilled water for 30 min. Thereafter the samples were frozen and lyophilized. Iron content of these samples was determined using ICP-MS measurements (see below for details).
Compression tests
Compression testing was performed with the collagen scaffolds incorporated with USPIO-NH2. A collagen scaffold without USPIO incorporation was used as a control. Scaffolds measuring 13 mm diameter and 3 mm thickness were used. Testing was performed with dry scaffolds (n=5) or with scaffolds hydrated with phosphate buffered saline (PBS; pH 7.4) at room temperature (n=5). Compression testing was performed by placing the sample between two parallel plates. Samples were compressed with a speed of 10 mm/min, until a distance between the two plates of 0.5 mm was reached. This resulted in a final compression of the sample of 83%.
Magnetic resonance imaging
For MR measurements, the scaffolds were embedded in 10% (w/v) gelatine phantoms. Nuclear MR relaxometry of labeled scaffolds was performed in a clinical 3T whole-body MR scanner (Philips Achieva, Best, The Netherlands) using a knee coil (SENSE-flex-M; Philips, Best, The Netherlands) at room temperature. Transverse (T2) relaxation times were measured in 2D scan mode using a multi-slice, multi-shot spin-echo sequences with a 90° excitation pulse followed by a train of equally spaced 180° refocusing pulses [TR = 1500 ms, TE = 8 - 168 ms, number of echoes = 20, inter-echo spacing = 8 ms, reconstruction matrix = 124, voxel size = 1.7 × 1.7, slice thickness = 2 mm, scan mode = multi-shot (MS)]. For T2* relaxometry, images at 32 echo times (TE range = 3 - 99 ms) were acquired by using a multi-shot, multi-slice fast-field gradient-echo sequence [TR = 196 ms, 3 ms interval between two echoes, reconstruction matrix = 172, voxel size = 0.5 × 0.5 mm, slice thickness = 2 mm, 30° flip angle].
T2 and T2* relaxation times (R2 and R2*) were calculated by fitting an exponential curve to the signal amplitudes as a function of the echo time (TE) for each segmented scaffold region using the Imalytics Preclinical Software (Philips Technologie GmbH, Aachen, Germany). The exponential curve includes an offset to account for a signal plateau created by noise or a component with slow signal decay. Furthermore, T1- and T2-weighted images were acquired using a T1-weighted turbo-spin-echo (TSE) sequence [TR = 9 ms, TE = 700 ms, 0.164 × 0.164 mm voxel size, slice thickness = 2 mm], and a T2-weighted TSE sequence [TR = 1200 ms, TE = 100 ms, 0.2 × 0.2 mm voxel size, slice thickness = 2 mm]. T2* gradient-echo image datasets were used to prepare Susceptibility Gradient Mapping (SGM) images using the MRI Positive Contrast Tool V 3.0 software (Philips, Best, The Netherlands). The size and volume of the scaffolds were assessed based on T1- and T2-weighted TSE images using the Imalytics Preclinical Software (Philips Technologie GmbH, Aachen, Germany).
Prussian Blue Staining
Prussian Blue staining was performed to validate the localization of USPIO nanoparticles in the scaffolds. To this end, fixed scaffold sections were first incubated with a 10% aqueous solution of K4Fe(CN)6 for 5 min, and then with a 1:1 mixture of 10% K4Fe(CN)6 and 20% HCl for 30 min. After staining, the scaffold sections were washed with water and mounted with coverslips for microscopy analysis (Zeiss Axio Imager M2).
Inductively Coupled Plasma Mass Spectroscopy Analysis
Digestion of the scaffolds was performed in a closed vessel microwave reaction system (MLS ethos plus, MPV-100/HAT) after addition of 1.5 mL nitric acid (65%) to 1.5 mL hydrogen peroxide and 1 mL of internal standard (rhodium) using 1000 W for 45 min by raising the temperature from 25 °C to 210 °C and left constant at 210°C for another 15 min. Samples were diluted 1:100 in water. The amount of incorporated iron was determined using a high-resolution sector field inductively coupled plasma mass spectrometer (ICP-MS; Elan-DRCII, Perkin Elmer) equipped with an injector (2.0 mm i.d.). Data were acquired at medium resolution. A Quartz Cyclonic spray chamber equipped with a Meinhard Type A quartz nebulizer was used for sample introduction. The iron content was expressed in grams of iron per kg of scaffold material.
Collagenase Assay
Scaffolds were weighed and transferred into 1 mL TRIS-HCl (1 M; pH 7.4). Five units/mg of collagenase were added, and the scaffolds were incubated with the enzyme for 1-5 days at 37 °C. After stopping enzymatic degradation by the addition of 1 mL of 0.01 M EDTA, the scaffolds were embedded in 10% (w/v) gelatine and imaged using a T2-weighted TSE sequence [TR = 100 ms, TE = 1200 ms, 0.164 × 0.164 mm voxel size, slice thickness = 2 mm].
Cell culture
NIH3T3 and SMC were cultured in DMEM (Gibco, Invitrogen, Germany) cell culture medium supplemented with 10% fetal calf serum (FCS; Invitrogen, Germany) and 1% Pen/Strep (10000 U/mL penicillin; 10000 mg/mL streptomycin, Invitrogen, Germany). HUVEC were cultured using human endothelial cell growth medium (VascuLife, Promocell, Germany) containing 2% growth supplements, 3% FCS and 1% Pen/Strep. Cells were cultured in T75 cell culture flasks (Cell Star, Greiner, Germany) and incubated at 37 °C, 5% CO2 and 95% relative humidity.
Scaffold Colonization, Staining and Microscopy
Cells were diluted to a concentration of 5000 cells/μl. The scaffolds (13 mm in diameter, 3 mm in height) had a fluid capacity of 97% (v/v), corresponding to ~400 μl. For cell seeding, 400 μl of cell suspension were dropped onto the scaffold, resulting in the seeding of 2*106 cells per scaffold (i.e. 1.5*106 cells per cm2). After 1 and 8 days of colonization, the scaffolds were embedded in Tissue-Tek, frozen in liquid nitrogen vapor and cut in 7-10 μm slices. Sections were fixed for 5 min in 80% methanol at 4°C followed by 2 min in acetone at 20°C, and then rehydrated in PBS. Stainings and quantitative microscopy analyses were conducted for scaffolds colonized with NIH3T3, HUVEC and SMC. H&E stainings were done according to standard histopathological protocols. Immunofluorescence stainings were performed using a primary antibody against the proliferation marker Ki67, which was incubated for 60 min at room temperature, followed by secondary antibody staining and nuclear counterstaining using Hoechst. Apoptotic cells were visualized by conducting a TUNEL assay (Roche, Germany). Stained sections were examined and photographed with the Zeiss Axio Imager M2. Images were fused to get a transversal cross section of the scaffolds and segmented in ten different sections of 0.3 × 0.3 mm. To evaluate the impact of USPIO-labeling on cellular colonization, the total number of cells as well as the number of proliferating Ki67-positive cells were quantified and compared. For quantifying proliferating and apoptotic cells in the scaffolds, 3 micrographs (100x magnification) from 3 representative sections per scaffold upon 1 and 8 days of colonization were analyzed for each condition. Area fractions of Ki67 and TUNEL-positive cells were determined. Quantifications were performed using the AxioVision Rel 4.8 software (Zeiss, Göttingen, Germany).
In vivo evaluation
All animal experiments were approved by the governmental review committee on animal care. Twelve B6 mice (Charles River Laboratories International, Inc., Wilmington, MA, USA) were used for the in vivo evaluation of 6 non-labeled and 6 labeled (0.7% (w/w) USPIO 3D) collagen patches. Patches were implanted subcutaneously into the right hind limb of the mice under isoflurane anesthesia. At day 1, 8, 15 and 22 post implantation, MR investigations of the implanted scaffolds were performed in three mice, and one mouse from each group was sacrificed to remove the scaffold for ex vivo histological analysis and macrophage staining. MR imaging of the implanted scaffold was carried out under isoflurane anesthesia using a clinical 3T MR scanner, in combination with a custom-made small animal solenoid sense-receive mouse coil (40 mm inner diameter and 78 mm bore length; resulting in a 40 mm field of view) (Philips Research Laboratories, Hamburg, Germany). The head-to-tail symmetry line was placed perpendicular to B0 in the magnet bore. To visualize anatomic details, transverse high-spatial resolution T1 and T2-weighted MR images of the scaffolds were acquired by using a multi-shot turbo-spin-echo sequence with the following parameters: T1 [TR = 500 ms, TE = 20 ms, FA = 90°, 0.3 × 0.3 mm voxel size, slice thickness = 1 mm, reconstruction matrix = 192]; T2 [TR = 1200 ms, TE = 100 ms, FA = 90°, 0.2 × 0.2 mm voxel size, slice thickness = 0.5 mm, reconstruction matrix = 256]. For relaxometry measurements T2 and T2*-weighted MR sequences were conducted: T2 [TR = 1500 ms, TE = 26 -340 ms, FA = 90°, refocusing pulse: 180°, number of echoes = 10, inter-echo spacing = 8 ms, reconstruction matrix = 224, voxel size = 0.4 × 0.4, slice thickness = 1 mm, scan mode = fast field echo (FFE)], T2*[ TR = 557 ms, TE =14 -171, FA = 30°, number of echoes = 15, inter-echo spacing = 11 ms, reconstruction matrix = 192, voxel size = 0.25 × 0.25, slice thickness = 1 mm, scan mode = fast field echo (FFE)].
Ex vivo analysis
H&E stainings were done according to standard histopathological protocols. Immunofluorescence stainings were performed using a primary antibody rat anti-mouse F4/80 (AbD Serotec) (1:50) in 12% BSA/PBS, which was incubated for 60 min at room temperature, followed by secondary antibody staining (Donkey ani-rat Cy3 (Dianova) (1:500) in 12% BSA/PBS 45 min at RT) and nuclear counterstaining using Hoechst. Stained sections were examined and photographed with the Zeiss Axio Imager M2. For quantifying the infiltration of macrophages in the scaffolds, 3 micrographs (100x magnification) from 3 representative sections per scaffold from day 1, 8, 15, 22 were analyzed for each condition. Area fractions of F4/80-positive cells were determined. Quantifications were performed using the AxioVision Rel 4.8 software (Zeiss, Göttingen, Germany).
Statistical Analysis
Correlation coefficients (Pearson) and P values (two-tailed) between R2 and iron concentration were calculated for each scaffold type using the GraphPad Software (version 4.0). P<0.05 was considered to represent statistical significance. For the comparison of the mechanical stress values and for IHC area fraction analysis, one-way ANOVA with Bonferroni post-hoc correction was performed.
Supplementary Material
Acknowledgements
This work was supported by NRW / EU-Ziel 2-Programm (EFRE) 2007-2013: “Entwicklung und Bildgebung patientenoptimierter Implantate”, by the European Research Council (ERC Starting Grant 309495: NeoNaNo), and by the Portfolio grant (Helmholtz-Society) “Technologie und Medizin – Multimodale Bildgebung zur Aufklärung des In-vivo-Verhaltens von polymeren Biomaterialien”. The authors kindly acknowledge Manfred Bovi (Electron Microscopic Facility, University Clinic, RWTH-Aachen University) for assistance with the SEM measurements, Anne Rix for assistance with the MRI measurements and Susanne Golombek for assistance with the in vitro studies.
Contributor Information
Marianne E. Mertens, Department for Experimental Molecular Imaging University Clinic and Helmholtz Institute for Biomedical Engineering RWTH - Aachen University Pauwelsstrasse 20, 52074 Aachen (Germany)
Alina Hermann, Matricel GmbH Kaiserstraße 100 52134 Herzogenrath (Germany).
Anne Bühren, Matricel GmbH Kaiserstraße 100 52134 Herzogenrath (Germany).
Leon Olde-Damink, Matricel GmbH Kaiserstraße 100 52134 Herzogenrath (Germany).
Diana Möckel, Department for Experimental Molecular Imaging University Clinic and Helmholtz Institute for Biomedical Engineering RWTH - Aachen University Pauwelsstrasse 20, 52074 Aachen (Germany).
Felix Gremse, Department for Experimental Molecular Imaging University Clinic and Helmholtz Institute for Biomedical Engineering RWTH - Aachen University Pauwelsstrasse 20, 52074 Aachen (Germany).
Dr. Josef Ehling, Department for Experimental Molecular Imaging University Clinic and Helmholtz Institute for Biomedical Engineering RWTH - Aachen University Pauwelsstrasse 20, 52074 Aachen (Germany)
Prof. Dr. Fabian Kiessling, Department for Experimental Molecular Imaging University Clinic and Helmholtz Institute for Biomedical Engineering RWTH - Aachen University Pauwelsstrasse 20, 52074 Aachen (Germany)
Dr. Twan Lammers, Department for Experimental Molecular Imaging University Clinic and Helmholtz Institute for Biomedical Engineering RWTH - Aachen University Pauwelsstrasse 20, 52074 Aachen (Germany); Department of Controlled Drug Delivery MIRA Institute for Biomedical Engineering and Technical Medicine University of Twente PO Box 217, 7500 AE Enschede (The Netherlands).
References
- [1].Place ES, Evans ND, Stevens MM. Nat Mater. 2009;8:457–470. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- [2].Shi J, Votruba AR, Farokhzad OC, Langer R. Nano Lett. 2010;10:3223–3230. doi: 10.1021/nl102184c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hutmacher DW. Biomaterials. 2000;21:2529–2543. doi: 10.1016/s0142-9612(00)00121-6. [DOI] [PubMed] [Google Scholar]
- [4].Ma H, Hu J, Ma PX. Adv Funct Mater. 2010;20:2833–2841. doi: 10.1002/adfm.201000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wei G, Ma PX. Adv Funct Mater. 2008;18:3568–3582. doi: 10.1002/adfm.200800662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pashuck ET, Stevens MM. Sci Transl Med. 2012;4:160sr4. doi: 10.1126/scitranslmed.3002717. [DOI] [PubMed] [Google Scholar]
- [7].Griffith LG. Science. 2002;295:1009–1014. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- [8].L’Heureux N, Dusserre N, Marini A, Garrido S, Fuente L. d. l., McAllister T. Nat Clin Pract Cardiovasc Med. 2007;4:389–395. doi: 10.1038/ncpcardio0930. [DOI] [PubMed] [Google Scholar]
- [9].Bulte JWM, Kraitchman DL. NMR in Biomed. 2004;17:484–499. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- [10].Cheong S, Ferguson P, Feindel KW, Hermans IF, Callaghan PT, Meyer C, Slocombe A, Su CH, Cheng FY, Yeh CS, Ingham B, Toney MF, Tilley RD. Angew Chem Int Ed Engl. 2011;123:4206–4209. doi: 10.1002/anie.201100562. [DOI] [PubMed] [Google Scholar]
- [11].Cen L, Neoh KG, Sun J, Hu F, Liu W, Cui L, Cao Y. Adv Funct Mater. 2009;19:1158–1166. [Google Scholar]
- [12].Nelson GN, Roh JD, Mirensky TL, Wang Y, Yi T, Tellides G, Pober JS, Shkarin P, Shapiro EM, Saltzman WM, Papademetris X, Fahmy TM, Breuer CK. FASEB J. 2008;22:3888–3895. doi: 10.1096/fj.08-107367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lyons FG, Al-Munajjed AA, Kieran SM, Toner ME, Murphy CM, Duffy GP, O’Brien FJ. Biomaterials. 2010;31:9232–9243. doi: 10.1016/j.biomaterials.2010.08.056. [DOI] [PubMed] [Google Scholar]
- [14].Cheema U, Chuo CB, Sarathchandra P, Nazhat SN, Brown RA. Adv Funct Mater. 2007;17:2426–2431. [Google Scholar]
- [15].Köse GT, Korkusuz F, Özkul A, Soysal Y, Ödemir T, Yildiz C, Hasirci V. Biomaterials. 2005;26:5187–5197. doi: 10.1016/j.biomaterials.2005.01.037. [DOI] [PubMed] [Google Scholar]
- [16].Glowacki J, Mizuno S. Biopolymers. 2008;89:338–344. doi: 10.1002/bip.20871. [DOI] [PubMed] [Google Scholar]
- [17].Lu Q, Ganesan K, Simionescu DT, Vyavahare NR. Biomaterials. 2004;25:5227–5237. doi: 10.1016/j.biomaterials.2003.12.019. [DOI] [PubMed] [Google Scholar]
- [18].Freed LE, Vunjak-Novakovic G, Biron RJ, Eagles DB, Lesnoy DC, Barlow SK, Langer R. Nat Biotechnol. 1994;12:689–693. doi: 10.1038/nbt0794-689. [DOI] [PubMed] [Google Scholar]
- [19].Chan TR, Stahl PJ, Yu SM. Adv Funct Mater. 2011;21:4252–4262. doi: 10.1002/adfm.201101163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Schoof H, Apel J, Heschel I, Rau G. J Biomed Mater Res. 2001;58:352–357. doi: 10.1002/jbm.1028. [DOI] [PubMed] [Google Scholar]
- [21].Olde Damink LH, Dijkstra PJ, van Luyn MJ, van Wachem PB, Nieuwenhuis P, Feijen J. Biomaterials. 1996;17:765–773. doi: 10.1016/0142-9612(96)81413-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.