Figure 3. The miRNAs of Amphimedon queenslandica.
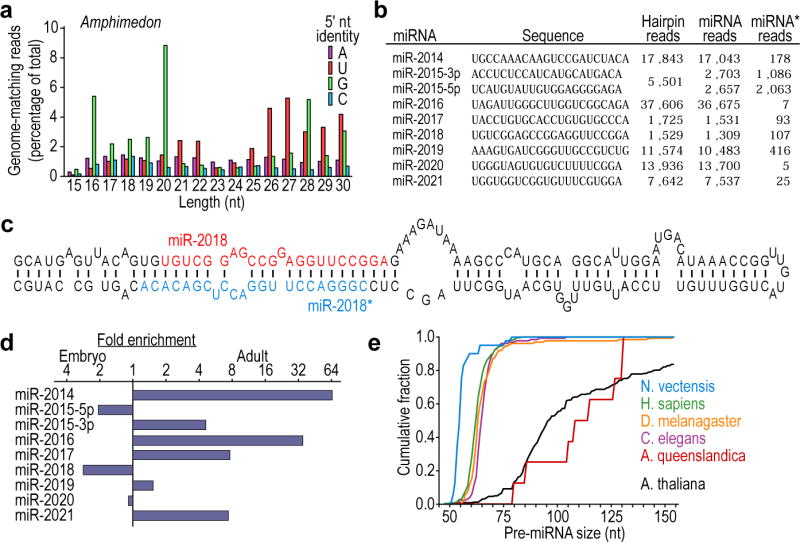
a, Length distribution of genome-matching sequencing reads representing small RNAs, plotted by 5′-nucleotide identity. Matches to ribosomal DNA were omitted. b, The Amphimedon miRNAs, shown as in Fig. 2d. Information analogous to that of Fig. 2b is provided for these miRNAs (Supplementary Data 2). c, Predicted secondary structure of the mir-2018 hairpin. d, Relative expression of Amphimedon miRNAs, as indicated by sequencing frequency from adult and embryo samples. e, Cumulative distributions of pre-miRNA lengths from miRNA transcripts of the species indicated. Amphimedon pre-miRNAs were significantly larger than those from any other animal species examined (P < 10−5, Wilcoxon rank-sum test), whereas those from Nematostella were significantly smaller (P < 10−5).
