Abstract
Analysis of 27 rotavirus strains from vaccinated and unvaccinated children revealed reassortment events in 3 strains: a gene derived from a vaccine; a gene acquired from a circulating strain; and reassortment between circulating strains. Data suggest that the widespread use of this monovalent rotavirus vaccine may introduce vaccine genes into circulating human rotaviruses or vice versa.
Keywords: Rotarix, vaccine, G1P[8] strains, phylogenetic analysis, reassortment, rotavirus, RVA, viruses, Brazil
Group A rotaviruses (RVAs) are a frequent cause of diarrhea in children. The RVA genome consists of 11 dsRNA segments that encode 6 structural (VP1–VP4, VP6, VP7) and 6 non-structural (NSP1– NSP6) proteins (1). The basis of a new classification system is phylogenetic analysis of 11 RVA genome segments, although binary classification is still used; genotyping is based on the coding genes for VP7 (G) and VP4 (P) (2).
Vaccination is considered effective in reducing the consequences of RVA. Two vaccines, Rotarix (Glaxo SmithKline, Brentford, UK) and RotaTeq (Merck & Co., Whitehouse Station, NJ, USA), are licensed in several countries. Both vaccines demonstrated broad protection against the most common RVA genotypes (3).
In Brazil, Rotarix, a monovalent attenuated human rotavirus vaccine for infants 6–24 weeks of age, was introduced in the National Immunization Programs in March, 2006. The vaccine is delivered in 2 doses, >4 weeks apart. In 2009, when vaccine coverage achieved 85.9%, reduction in hospitalization associated with diarrhea (17%) and in related mortality rates (22%) was observed (4). This study aimed to assess phylogenetic relationships between G1P[8] RVA obtained from vaccinated and unvaccinated children hospitalized for acute gastroenteritis.
The Study
During 2008–2010, fecal samples were collected from 3,852 children; 702 specimens (18.2%) had RVA-positive ELISA results; 27 of those (3.8%) were characterized as G1P[8] by using reverse transcription-PCR. Eighteen of the 27 specimens were from vaccinated children (Table 1). Study methods were approved by Fiocruz Ethical Committee (No. 311/06).
Table 1. Clinical features of 27 patients infected with group A rotaviruses (G1P[8]), Brazil 2008–2010*.
| Strain | State | Age (mo)† | Rotarix doses (mo/yr) |
Time elapsed between vaccination and hospitalization | Clinical symptoms |
||||
|---|---|---|---|---|---|---|---|---|---|
| 1st | 2nd | Diarrhea‡ | Blood | Fever | Vomit | ||||
| ES15221–08 | ES | 42 | – | – | – | – | – | – | – |
| SE15901–08 | SE | 2 | 09/2008 | – | 7 d | 3 | No | No | Yes |
| SE 16536–09 | SE | 2 | – | – | – | 2 | No | Yes | Yes |
| SE 16537–09 | SE | 4 | 03/2009 | – | 2 mo | 6 | No | Yes | Yes |
| SE 16779–09 | SE | 41 | 03/2006 | 05/2006 | 37 mo | 1 | Yes | Yes | Yes |
| SE 16782–09 | SE | 19 | 02/2008 | 04/2008 | 15 mo | 3 | No | No | y |
| SE 16800–09 | SE | 7 | 02/2009 | – | 5 mo | 3 | – | No | No |
| SE 16803–09 | SE | 10 | 11/2008 | 01/2009 | 6 mo | 1 | No | Yes | Yes |
| SE 16894–09 | SE | 20 | 02/2008 | 05/2008 | 15 mo | 2 | No | Yes | No |
| SE 16897–09 | SE | 15 | 08/2008 | 10/2008 | 10 mo | 1 | No | No | Yes |
| SE 16898–09 | SE | 4 | 06/2009 | – | 2 mo | 1 | No | No | Yes |
| SE 16977–09 | SE | 9 | 02/2009 | 04/2009 | 4 mo | 13 | No | Yes | Yes |
| SE 16978–09 | SE | 10 | 01/2009 | 03/2009 | 6 mo | 2 | No | Yes | No |
| SE 17120–09 | SE | 22 | 02/2008 | 05/2008 | 17 mo | 1 | No | No | Yes |
| SE 17122–09 | SE | 9 | 07/2008 | 09/2008 | 5 mo | 2 | – | Yes | No |
| SE 17123–09 | SE | 8 | – | – | – | 3 | No | Yes | Yes |
| SE 17241–09 | SE | 7 | 06/2008 | 08/2008 | 3 mo | 2 | No | Yes | Yes |
| PE17887–09 | PE | 6 | – | – | – | – | No | Yes | Yes |
| PE 17888–09 | PE | 10 | 11/2008 | – | 8 mo | – | No | Yes | Yes |
| PE 17890–09 | PE | 20 | 02/2008 | 05/2008 | 18 mo | – | No | No | Yes |
| PE 17891–09 | PE | 12 | – | – | – | – | No | Yes | Yes |
| MA18999–10 | MA | 18 | – | – | – | – | – | – | – |
| MA 19006–10 | MA | 2 | 08/10 | – | 4 d | 1 | Yes | No | No |
| MA 19013–10 | MA | 10 | – | – | – | – | – | – | – |
| MA 19015–10 | MA | 48 | – | – | – | – | – | – | – |
| MA 19030–10 | MA | 32 | 03/2008 | – | 29 mo | 7 | Yes | No | No |
| BA19391–10 | BA | 2 | – | – | – | 2 | – | No | No |
*Prefixes represent origins of circulating strains in Brazil: ES, Espirito Santo; –, no data; SE, Sergipe; PE, Pernambuco; MA, Maranhão; BA, Bahia. †Age of child at time of fecal sample collection. ‡Number of days child had diarrhea before fecal sample collection.
RVA detection, genotyping, and sequencing were performed (5). Sequences were deposited in GenBank under accession numbers: JQ926436-JQ926600 and JX683535-JX683664. Sequences were compared with those in strains obtained from GenBank (including Rotarix JX943604.2–JX943614.2).
Strains analyzed belonged to the Wa-like genogroup (genotype 1); 26 strains showed a G1–P[8]–I1–R1–C1–M1–A1–N1–T1–E1–H1 genome constellation. One sample, collected from a child vaccinated with 1 dose during 2010 in Maranhão (MA) state (MA19030–10), contained the G1–P[8]–Ix–R1–Cx–M1–A1–N1–T3–E1–H1 constellation.
Nucleotide (nt) identity values between circulating strains in Brazil and the Rotarix strain ranged 76%–100% (Table 2). Three samples showed 100% nt identity with the Rotarix strain in >1 gene. The SE15901–08 strain, collected on day 7 after the first dose, showed 100% nt identity with all Rotarix strain gene segments and could represent vaccine shedding. Vaccine antigen excretion detected by ELISA achieved 80%, declining to 18%–24% when collected at day 30; 11%–16% of children shed the virus at day 45 (6). Shedding is not associated with increased gastroenteritis-like symptoms. Widespread use of Rotarix might reveal adverse reactions not observed in clinical trials, emphasizing the need for global surveillance (7).
Table 2. Nucleotide similarity values when comparing the Brazilian G1P[8] samples with the Rotarix strain gene segments and genotype constellation of selected human group A rotaviruses*.
| Protein encoded by genes† |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain names‡ | Genotype constellation | VP7 | VP4 | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 |
| SE15901–09 | Vaccine | G1 100% | P[8] 100% | I1 100% | R1 100% | C1 100% | M1 100% | A1 100% | N1 100% | T1 100% | E1 100% | H1 100% |
| MA19006–10 | Vaccine | G1 100% | P[8] 99% | I1 99% | R1 99% | C1 98% | M1 99% | A1 98% | N1 100% | T1 100% | E1 99% | H1 98% |
| BA19391–10 | Vaccine | G1 100% | P[8] 99% | I1 100% | R1 99% | C1 98% | M1 100% | A1 100% | N1 100% | T1 100% | E1 99,6% | H1 100% |
| ES15221–08 | Wa-like (ES) | G1 94% | P[8] 90% | I1 90% | R1 100% | C1 90% | M1 93% | A1 92% | N1 91% | T1 97% | E1 92% | H1 92% |
| SE16800–09 | Wa-like (SE) | G1 94% | P[8] 92% | I1 89% | R1 97% | C1 90% | M1 96% | A1 92% | N1 88% | T1 98% | E1 98% | H1 92% |
| PE17888–09 | Wa-like (PE) | G1 94% | P[8] 92% | I1 90% | R1 94% | C1 91% | M1 93% | A1 93% | N1 90% | T1 97% | E1 94% | H1 93% |
| MA19013–10 | Wa-like (MA) | G1 95% | P[8] 93% | I1 92% | R1 95% | C1 92% | M1 95% | A1 94% | N1 92% | T1 97% | E1 94% | H1 93% |
| MA19030–10 | Wa/AU-1 reassortant | G1 93% | P[8] 91% | X | R1 93% | X | M1 94% | A1 93% | N1 89% | T3 76% | E1 92% | H1 93% |
| Wa | Wa-like | G1 | P[8] | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| DS-1 | DS-1-like | G2 | P[4] | I2 | R2 | C2 | M2 | A2 | N2 | T2 | E2 | H2 |
| AK26 | Wa/DS-1 reassortant | G2 | P[4] | I2 | R2 | C2 | M2 | A2 | N1 | T2 | E2 | H2 |
| 6809 | Wa/DS-1 reassortant | G8 | P[6] | I2 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Matlab13 | Wa/DS-1 reassortant | G12 | P[6 | I1 | R1 | C1 | M1 | A1 | T2 | T1 | E1 | H1 |
| Mani-253 | Wa/DS-1 reassortant | G4 | P[4] | I1 | R1 | C1 | M2 | A8 | N1 | T1 | E1 | H1 |
| AU-1 | AU-1-like | G3 | P[9] | I3 | R3 | C3 | M3 | A3 | N3 | T3 | E3 | H3 |
| T152 | AU-1-like | G12 | P[9] | I3 | R3 | C3 | M3 | A12 | N3 | T3 | E3 | H6 |
| K8 | Wa/AU-1 reassortant | G1 | P[9] | I1 | R3 | C3 | M3 | A1 | N1 | T3 | E3 | H3 |
| Mani-265 | DS-1/AU-1 reassortant | G10 | P[6] | I2 | R2 | C2 | M2 | A3 | N2 | T7 | E2 | H2 |
*Wa-like, DS-1-like, AU-like genotypes are shown in green, red, and orange, respectively, and the P[6] genotype is shown in blue; X, indeterminate genotypes; –, no data. †One sample for each group was chosen as representative and the average was shown. ‡Prefixes represent origins of circulating strains in Brazil: SE, Sergipe; BA, Bahia; MA, Maranhão; ES, Espirito Santo; PE, Pernambuco.
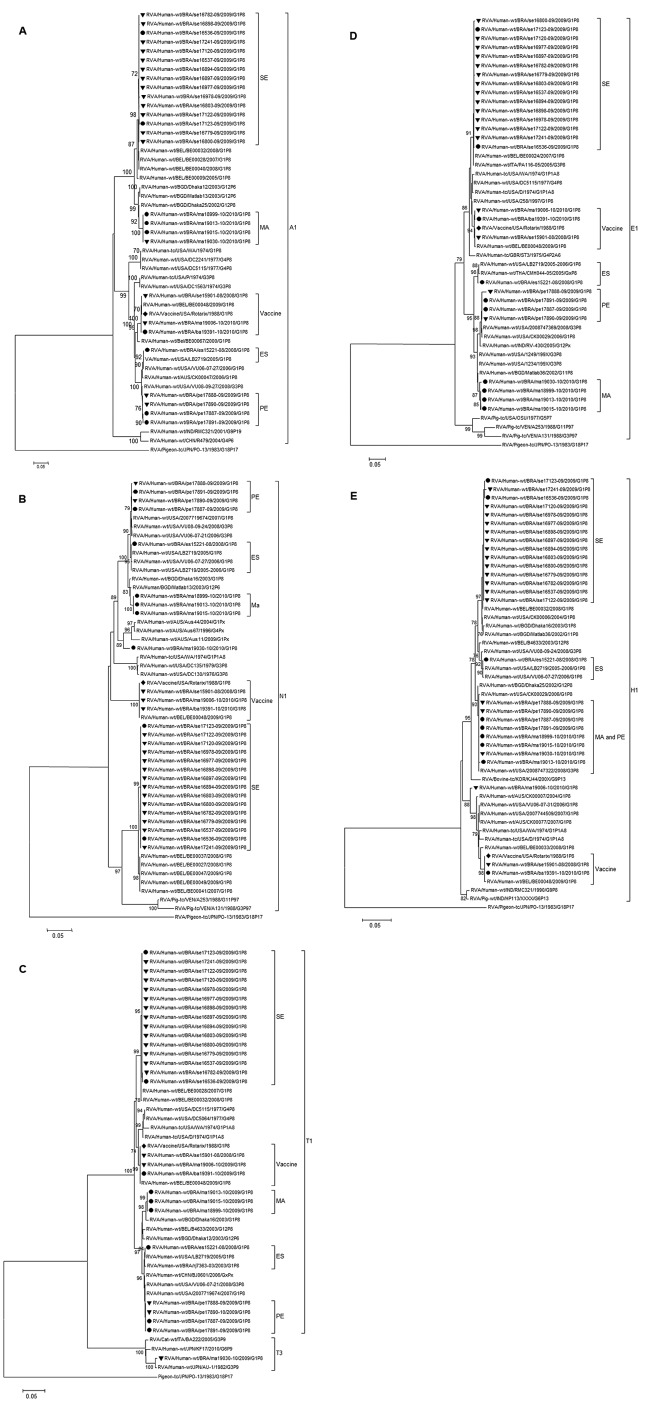
In phylogenetic analysis of 11 genes, 10 genes clustered into 5 clades. In the remaining gene, NSP5, the circulating strains clustered into 4 groups (Figure 1, panel E).
Figure 1.
Phylogenetic analysis of nucleotide sequence of A) nonstructural protein 1 [NSP1], B) NSP2, C), NSP3, D) NSP4, and E) NSP5 encoding genes of G1P[8] RVA samples collected in Brazil and described in this study. Filled circles indicate samples from non-vaccinated children and filled triangles vaccinated children. The Rotarix strain is indicated by a filled diamond. Bootstrap values >70%, estimated with 2,000 pseudoreplicate datasets, are indicated at each node. Only the genotype in which the strains under investigation cluster is shown completely; 1 prototype from another genotype has been added as an outgroup. Scale bars represent 0.5 substitutions per nucleotide site.
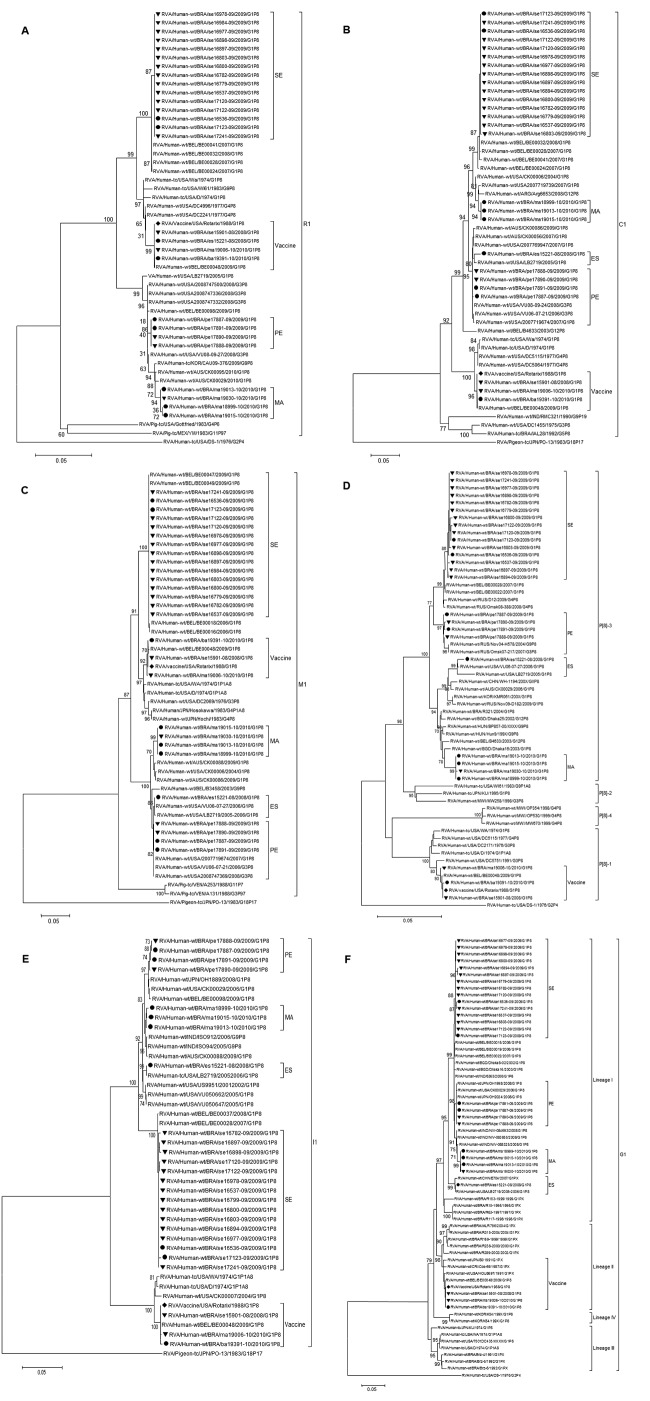
Strain ES15221–08, detected in an unvaccinated child, is genetically distinct and differs in origin from other G1P[8] circulating strains (Figures 1,2). The VP1 gene (648 bp, Figure 2, panel A) in this sample showed 100% nt identity with the Rotarix strain. Strain MA19006–10 was genetically similar to the strain; however, the NSP5 segment (Figure 1, panel E) was closely related to a G1P[8] strain from Australia (JF490152). These 2 strains appear to have been generated by reassortment with this vaccine strain.
Figure 2.
Phylogenetic analysis of nucleotide sequence of (A) VP1, (B) VP2, (C), VP3, (D) VP4, (E) VP6 and (F) VP7 genes of G1P[8] RVA samples collected in Brazil and described in this study. Filled circles indicate samples from non-vaccinated children and filled triangles vaccinated children. The Rotarix vaccine strain is indicated by a filled diamond. Bootstrap values above 70%, estimated with 2,000 pseudoreplicate datasets, are indicated at each node. Only the genotype in which the strains under investigation cluster is shown completely and one prototype from another genotype has been added as outgroup. Scale bars represent 0.5 substitutions per nucleotide site.
Strain MA19030–10, detected in a child after 1 vaccine dose, was closely related to the MA group, but the NSP2 segment differed in origin from other MA samples (Figure 1, panel B) because it clustered with other Wa-like strains. The NSP3 gene belonged to genotype 3 because it was 99.2% (nt) similar to the AU-1 prototype strain (DQ490535.1). These results suggested reassortment events between Wa-like and AU-1 like co–circulating strains.
NSP2 and NSP3 genes from sample MA19030–10 and NSP5 genes from sample MA19006–10 differed from each other and from those of their respective clusters. Samples MA19006–10 and MA19030–10 are genetically distinct and might have distinct evolutionary histories. Studies that included this genogrouping system were performed to prove the existence of inter-genogroup reassortment between human RVA genogroups or human and animal genogroups. The existence and effectiveness of heterogeneous genome constellations remains unclear, probably because it is caused by mechanisms that create protein sets that work better when kept together (8).
Phylogenetic analysis of the VP8* (aa 1–247) portion of VP4 encoding gene showed circulating strains (Figure 2, panel D) clustered into 2 lineages (P[8]-3 and P[8]-1). The alignment of the deduced aa sequences showed potential trypsin cleavage sites at arginine 240 and 246 conserved in all samples. All circulating strains contained 91.3%–100% identical aa residues to the Rotarix strain in VP8* antigenic epitopes; no changes were observed on epitopes 8–2 and 8–4.
Two VP7 lineages (G1-I and G1-II) were identified. Samples closely related to the Rotarix strain belonged to G1-II; remaining strains belonged to G1-I. Comparing the regions defined as antigenic epitopes (7–1 and 7–2) for VP7 protein, we found >2 epitopes (aa 94, 123, 148, 217) were not conserved among circulating strains compared with the Rotarix strain. The following amino acids were conserved in the non-VP7/non-VP4 segments of either the Rotarix strain or other strains analyzed: cysteine residues involved in disulfide bonds at positions 6, 8, 85, and 285 in NSP2 protein; the leucine, isoleucine, aspartic acid, methionine and glutamine at positions 275, 281, 288, and 295 in the NSP3 protein; the N-linked glycosylation sites at positions 8 and 18, and cysteine residues at positions 63 and 71 in NSP4 protein; and serine residues at positions 153, 155, 163, and 165 in NSP5 protein.
Conclusions
This report characterizes the complete genome of G1P[8] strains in Brazil. Phylogenetic analysis showed that sequences clustered consistently with the region of sample collection. Three strains circulating in Brazil were closely related to those in Rotarix; 1 of the 3 was 100% identical to Rotarix, likely representing shedding of this vaccine. Sequence analysis confirmed the presence of the Rotarix VP1–derived segment in 1 sample (ES15221–08), indicating an unreported reassortment event between the vaccine and a community strain.
The backbone of MA19030–10 sample was Wa-like, but the NSP3 segment exhibited a T3 genotype that was described for the AU-1 genogroup. This finding suggests that this strain derived its NSP3 gene from an AU-1-like strain through reassortment.
Changes in antigenic regions of VP4 and VP7 proteins have been associated with mutated RVA strains that spread (9,10). Comparison of the aa sequences of VP7 and VP8* of strains circulating in Brazil and the monovalent vaccine strain demonstrated that VP7 and VP8* of the circulating strains showed similar antigenic regions to those of the vaccine. No differences between strains from vaccinated and unvaccinated children were observed.
Considering the segmented RVA genome and that the Rotarix vaccine is an attenuated RVA human strain, it is expected that reassortants will arise and circulate among humans. The effects of such events are not known. This study described strains that originated from reassortment events between the Rotarix vaccine strain and strains detected in vaccinated and unvaccinated children. Improvement of RVA surveillance programs that include full genome sequencing analysis will strengthen the understanding of how vaccines will affect the RVAs circulating among humans, and how those events could affect the use of live vaccines, the frequency of RVA intra- and intergenogroup reassortment events under natural conditions, and the stability of RVA generated by such events.
Acknowledgments
The authors thank the PDTIS DNA Sequence Platform (RPT01A) and LATER/BioManguinhos, FIOCRUZ, Rio de Janeiro, Brazil; and the Secretary of Public Health State of Sergipe, Maranhão, Espírito Santo, Bahia and Pernambuco. We also thank Alexandre Madi Fialho and Rosane Maria Santos de Assis for technical assistance.
This research was supported by federal funds from the Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES), the National Council for Scientific and Technological Development (CNPq), project PAPES VI/Fiocruz - CNPq, Oswaldo Cruz Institute (PROEP – Fiocruz - CNPq), the General Coordination of Public Health Laboratories – Secretary of Health Surveillance (CGLAB/SVS), project CAPES-MERCOSUL PPCP 023/2011, and Carlos Chagas Filho Foundation for Research Support of Rio de Janeiro State (FAPERJ). T.L.R. receives a post-doctoral scholarship from CNPq.
Biography
Dr Rose is a post-doctoral student at the Laboratory of Comparative and Environmental Virology, Oswaldo Cruz Institute, Fiocruz, Brazil. Her research interests include RVA molecular epidemiology.
Footnotes
Suggested citation for this article: (Rose TL, Silva MFM, Gomez MM, Resque HR, Ichihara MYT, Volotão EM, et al. Evidence of Vaccine-related Reassortment of Rotavirus, Brazil, 2008–2010. Emerg Infect Dis. 2013 Nov [date cited]. http://dx.doi.org/10.3201/eid1911.121407
These authors contributed equally to this article.
References
- 1.Estes MK, Kapikian AZ. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Martin MA, Lamb RA, Roizman B, et al., editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2007. p. 1917–74. [Google Scholar]
- 2.Zeller M, Patton JT, Heylen E, de Coster S, Ciarlet M, Van Ranst M, et al. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J Clin Microbiol. 2012;50:966–76. PubMed 10.1128/JCM.05590-11 [DOI] [PMC free article] [PubMed]
- 3.Patton JT. Rotavirus diversity and evolution in the post-vaccine world. [PubMed]. Discov Med. 2012;13:85–97. [PMC free article] [PubMed] [Google Scholar]
- 4.do Carmo GMI, Yen C, Cortes J, Siqueira AA, de Oliveira WK, Cortez-Escalante JJ, et al. Decline in diarrhea mortality and admissions after routine childhood rotavirus immunization in Brazil: a time-series analysis. PLoS Med. 2011;8:e1001024. PubMed 10.1371/journal.pmed.1001024 [DOI] [PMC free article] [PubMed]
- 5.Gómez MM, da Silva MFM, Zeller M, Heylen E, Matthijnssens J, Ichihara MYT, et al. Phylogenetic analysis of G1P[6] group A rotavirus strains detected in Northeast Brazilian children fully vaccinated with Rotarix. Infect Genet Evol. PubMed. 2013. In press . 10.1016/j.meegid.2013.03.028 [DOI] [PubMed] [Google Scholar]
- 6.Phua KB, Quak SH, Lee BW, Emmanuel SC, Goh P, Han HH, et al. Evaluation of RIX4414, a live, attenuated rotavirus vaccine, in a randomized, double-blind, placebo-controlled phase 2 trial involving 2464 Singaporean infants. J Infect Dis. 2005;192(Suppl 1):S6–16. PubMed 10.1086/431511 [DOI] [PubMed]
- 7.O’Ryan M, Linhares AC. Update on RotarixTM: an oral human rotavirus vaccine. Expert Rev Vaccines. 2009;8:1627–41. PubMed 10.1586/erv.09.136 [DOI] [PubMed]
- 8.Heiman EM, McDonald SM, Barro M, Taraporewala ZF, Bar-Magen T, Patton JT, et al. Human Rotavirus Genomics: Evidence that gene constellations are influenced by viral protein interactions. J Virol. 2008;82:11106–16. PubMed 10.1128/JVI.01402-08 [DOI] [PMC free article] [PubMed]
- 9.Diwakarla CS, Palombo EA. Genetic and antigenic variation of capsid protein VP7 of serotype G1 human rotavirus isolates. [PubMed]. J Gen Virol. 1999;80:341–4. [DOI] [PubMed] [Google Scholar]
- 10.Dyall-Smith ML, Lazdins I, Tregear GW, Holmes IH. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc Natl Acad Sci U S A. 1986;83:3465–8. PubMed 10.1073/pnas.83.10.3465 [DOI] [PMC free article] [PubMed]