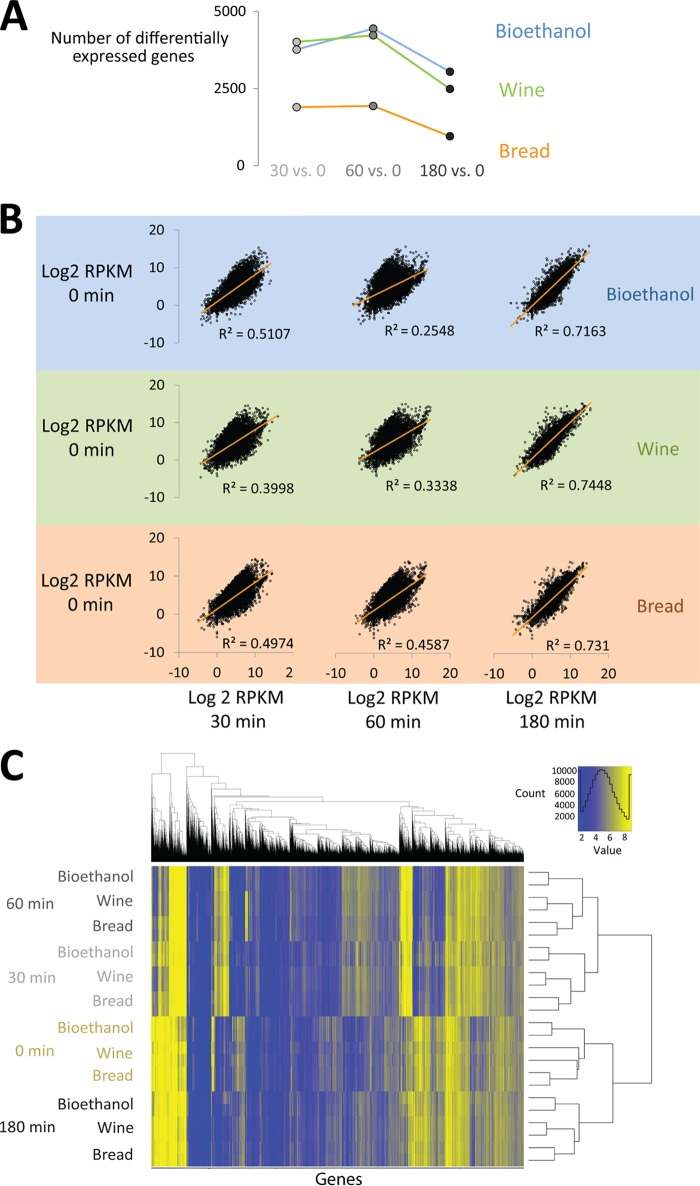
Fig 2.
(A) A large number of genes are differentially expressed at different time points during bread dough fermentation compared to the 0-min sample. The commercial baker's yeast (“Bread”) strain induces fewer changes in the transcriptome in response to the new environment. Note that differentially expressed genes include both protein- and RNA-coding genes. (B) The transcriptome of cells at the latest fermentation time point shows the highest level of similarity to cells in stationary phase (0-min sample). Shown are pairwise scatter plots and correlation coefficients for all mRNA quantities (as estimated by RPKM values) of the nonfermenting sample (0 min) and the three fermentation time points (30, 60, and 180 min) for the three different yeast strains (bioethanol, wine, and bread yeast). Note that the correlation between biological replicates was very high (R2 of between 0.959 and 0.997) (data not shown). (C) Clustering of expression profiles shows that the different yeast strains display largely similar (but not identical) gene expression patterns during bread dough fermentation. The graph shows hierarchical clustering of all expression data (including biological replicates for each strain and each time point). Every horizontal line contains the expression data (RPKM values) for a given strain and sampling time (i.e., one sample). The horizontal lines are ordered based on their overall similarity, and the clustering tree on the right shows the similarity between the samples (see Materials and Methods).