Fig 1.
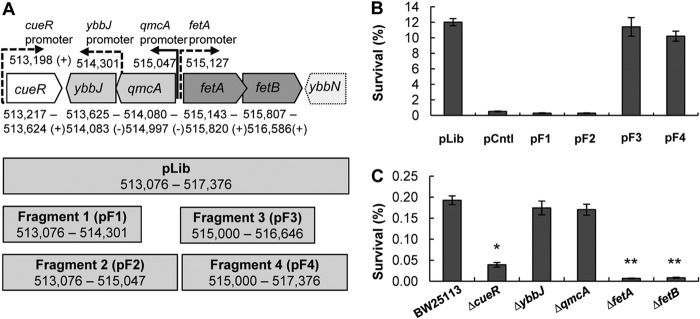
Role of the E. coli locus at positions 513217 to 517376 in H2O2 tolerance. (A, top) Genes included in plasmid pLib that enhances resistance to H2O2. pLib spans chromosomal positions 513217 to 517376 and includes the complete open reading frames of cueR, ybbJ, qmcA, fetA (ybbL), and fetB (ybbM) (solid lines) as well as the incomplete ybbN gene (dashed lines). Gene orientations and promoters (solid for experimental and dashed for predicted) were obtained from EcoCyc (45). (Bottom) Four fragments were cloned into plasmids pF1 to pF4 based on the sequence included in pLib. (B) Survival rates of E. coli BW25113 with plasmids pF1 to pF4 after a 30-min exposure to 4 mM H2O2 at 37°C. Cultures were grown to an OD600 of 1, diluted 10-fold in 5 ml of prewarmed LB medium, and stressed with H2O2 at 37°C for 30 min. Cell survival was determined by plating dilution series pre- and poststress as follows: % survival = [(CFU/ml)poststress/(CFU/ml)prestress] × 100. pLib was the plasmid isolated during library enrichment. pF1, pF2, pF3, and pF4 are plasmids with inserts that correspond to fragments 1 to 4 illustrated in panel A. Data are means ± standard errors of the means (n = 4). (C) Rates of survival of E. coli strain BW25113 and the ΔcueR, ΔybbJ, ΔqmcA, ΔfetA, and ΔfetB mutant strains against 4 mM H2O2 stress. Assays were performed as described above for panel B. Data are means ± standard errors of the means (n = 3). ∗ indicates a statistical difference (P value of <0.05) for a Student t test performed between E. coli strain BW25113 and the ΔcueR strain. ∗∗ indicates a statistical difference (P value of <0.05) for a Student t test performed between the ΔcueR and the ΔfetA or ΔfetB strains.
