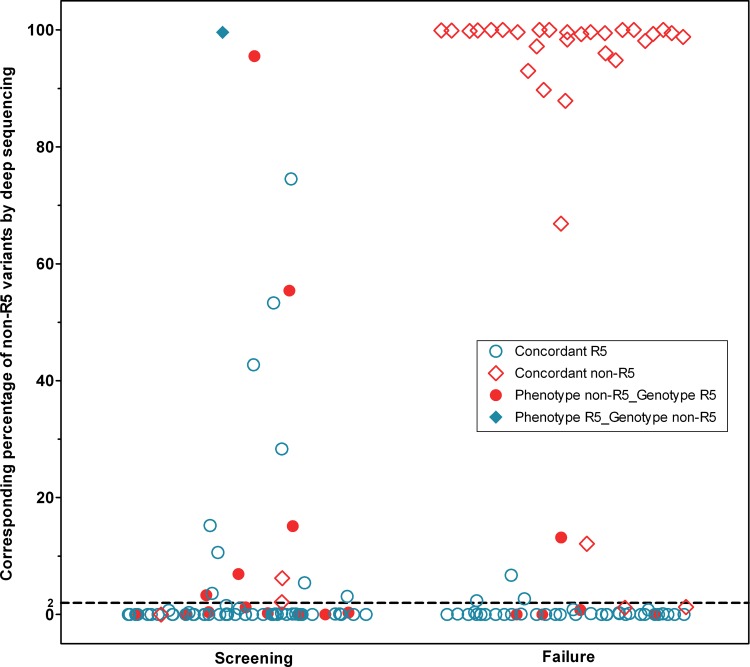
Fig 2.
Percentages of non-R5 variants by deep sequencing results at screening and failure. Shown is a scatter plot of the percent non-R5 variants for all patients with deep sequencing results at screening and failure. Points are marked by whether the tropism results for the same time point were concordant between phenotype and population-based genotype determinations (see key). The failure column illustrates how the majority of failure samples had very high or very low non-R5 prevalence. Determination of phenotypes was performed by ESTA at screening and by the original Trofile assay (OTA) at failure. A dashed line at a 2% non-R5 prevalence represents a cutoff for deep sequencing, above which a sample was classified as having non-R5 tropism.